Abstract
Tuberculosis patients may be infected with or have disease caused by more than one Mycobacterium tuberculosis strain, usually referred to as “mixed infections.” These have mainly been observed in settings with a very high tuberculosis incidence and/or high HIV prevalence. We assessed the rate of mixed infections in a population-based study in rural Vietnam, where the prevalences of both HIV and tuberculosis are substantially lower than those in previous studies looking at mixed infections. In total, 1,248 M. tuberculosis isolates from the same number of patients were subjected to IS6110 restriction fragment length polymorphism (RFLP) typing, spoligotyping, and variable-number-tandem-repeat (VNTR) typing. We compared mixed infections identified by the presence of (i) discrepant RFLP and spoligotype patterns in isolates from the same patient and (ii) double alleles at ≥2 loci by VNTR typing and assessed epidemiological characteristics of these infections. RFLP/spoligotyping and VNTR typing identified 39 (3.1%) and 60 (4.8%) mixed infections, respectively (Cohen's kappa statistic, 0.57). The number of loci with double alleles in the VNTR pattern was strongly associated with the proportion of isolates with mixed infections according to RFLP/spoligotyping (P < 0.001). Mixed infections occurred more frequently in newly treated than in previously treated patients, were significantly associated with minor X-ray abnormalities, and were almost significantly associated with lower sputum smear grades. Although the infection pressure in our study area is lower than that in previously studied populations, mixed M. tuberculosis infections do occur in rural South Vietnam in at least 3.1% of cases.
INTRODUCTION
For a long time it was assumed that a tuberculosis (TB) infection protects against a subsequent infection. In fact, vaccination against infectious diseases is based on this principle. However, in 1976 there were already anecdotal indications that TB patients can be reinfected by another Mycobacterium tuberculosis strain and that infections with multiple strains exist. Using phage typing, Bates et al. (2) found different phage types of M. tuberculosis in single hosts. The occurrence of infections with multiple M. tuberculosis strains was confirmed by using DNA fingerprinting techniques, first at the turn of the century in selected patients by Yeh et al. (25) and by Braden et al. (3) and then more recently in larger patient populations (14, 15, 22). The introduction of molecular techniques offered new possibilities for studying the natural history of TB infection more extensively. PCR assays targeting particular predominant M. tuberculosis genotype families were developed, and by applying these methods to clinical material, Warren et al. found that the rate of occurrence of “mixed infections,” i.e., infections with multiple M. tuberculosis strains, amounted to 19% of examined patients in South Africa (22).
High rates of mixed infections have been found in populations living in crowded conditions, including a high-density urban community (22) and a hospital (7) in South Africa and a prison in Georgia (15). However, the frequency of mixed infections in human populations with a lower tuberculosis infection pressure (e.g., populations under less crowded conditions and with a lower HIV prevalence) is unknown. Although the burden of TB is high in South Vietnam, with a prevalence of smear-positive TB of 219/100,000 population (95% confidence interval [CI], 145 to 294/100,000) (9), it is much lower than the prevalence in the studied areas in South Africa (1,000/100,000) (22). Similarly, the prevalence of HIV among TB patients is lower in Vietnam than in South Africa (8.2% versus 50 to 80%) (18; Brand South Africa Media Service).
We studied the occurrence of mixed infections in a population-based study in a rural area in South Vietnam using IS6110 restriction fragment length polymorphism (RFLP) typing and spoligotyping (11, 20). The former method is used to distinguish M. tuberculosis isolates at the strain level to study patient-to-patient transmission but also enables determination of the genotype to which the M. tuberculosis strain belongs, while the latter method can be used only for genotype determination (4). It is known from previous studies that about 35% of the M. tuberculosis isolates from South Vietnam are of the Beijing genotype, while about 49% of the isolates represent the East African Indian (EAI) genotype (previously known as the Vietnam genotype) (1, 5). The predominance of these two M. tuberculosis genotype families in South Vietnam and their highly characteristic IS6110 RFLP and spoligotype patterns enabled us to detect possible mixed infections with strains of these different genotypes in a reliable way by comparing the results of both typing methods. In addition, mixed infections were detected by the visualization of double alleles in variable-number-tandem-repeat (VNTR) typing patterns (13, 15). In this study, we were able to compare the sensitivity of the various typing approaches to detect mixed infections; however, because much is unknown about the evolution of VNTR patterns, we used RFLP/spoligotype results to quantify the occurrence of mixed infections and investigate possible risk factors for mixed infections. This is the first report of mixed infections in rural Vietnam, where the population density and HIV infection prevalence are low and TB incidence is moderate, in contrast to settings in previous studies looking at mixed infections.
MATERIALS AND METHODS
Patient population.
The study area consisted of three adjacent rural districts in Tien Giang Province, situated in the Mekong River Delta in southern Vietnam. All patients aged ≥15 years, resident in the study area, and registered for treatment of smear-positive pulmonary TB between 1 January 2003 and 31 December 2005 at the participating District Tuberculosis Units or at the provincial TB hospital were eligible for inclusion in the study (10). By interviews using prestructured questionnaires, we collected data on the sex, age, M. bovis BCG vaccination, X-ray abnormalities, educational level, marital status, occupation, and history of treatment of all participants. HIV testing was not done routinely. Treatment outcomes were based on routine smear examination (24), as described by Buu et al. (5). Buu et al. previously found that tuberculosis is usually transmitted outside the household in the study area (6); therefore, epidemiological links between patients were not assessed in in-depth interviews for the purpose of this study.
Mycobacterial isolates.
Sputum specimens were kept refrigerated and were transported to Pham Ngoc Thach Tuberculosis and Lung Disease Hospital in Ho Chi Minh City, Vietnam, within 72 h after collection. Specimens were decontaminated and liquefied with 1% N-acetylcysteine, 2% NaOH, inoculated on modified Ogawa medium, and incubated at 37°C. Cultures were examined for growth after 1, 2, 4, 6, and 8 weeks of incubation. Cultures with no growth after 8 weeks were reported to be negative. M. tuberculosis was identified using the niacin and nitrate tests (10).
Drug susceptibility testing.
Drug susceptibility testing was performed by the proportion method following the guidelines of the World Health Organization and the International Union against Tuberculosis and Lung Disease (23). Criteria for drug resistance were ≥1% colony growth (10) at 28 or 40 days compared to the growth on the drug-free control medium at the following drug concentrations: isoniazid, 0.2 μg/ml; rifampin, 40 μg/ml; streptomycin, 4 μg/ml; and ethambutol, 2 μg/ml. Multidrug resistance (MDR) was defined as resistance to both rifampin and isoniazid.
DNA typing.
We included all 1,248 patients (66% of the eligible population) with isolates on which all 3 typing methods were applied (RFLP, spoligo-, and VNTR typing). Genomic DNA was extracted from positive cultures by using a method described earlier (21). IS6110 RFLP typing and spoligotyping were performed according to the internationally standardized methods (11, 20). VNTR typing was done using 15 loci, as described by Supply et al. (17).
Definition of mixed infections by RFLP, spoligo-, and VNTR typing.
All isolates that yielded discrepant results with regard to M. tuberculosis genotype family in RFLP and spoligotyping were subjected to both typing methods for a second time from the same DNA to ensure the reproducibility of the observation. Beijing and EAI genotypes were assigned as reported elsewhere on the basis of the IS6110 RFLP and spoligotyping patterns (1, 4, 12). Isolates that repeatedly had a spoligotype characteristic of the EAI genotype and an IS6110 RFLP pattern characteristic of a Beijing genotype strain (12), or the other way around, or a spoligotype characteristic of the Beijing or EAI genotype and an IS6110 RFLP pattern characteristic of another genotype, or the other way around, were considered to represent a mixed infection.
VNTR typing has been shown to be sensitive in the detection of mixed infections by revealing double alleles, with mixed infections defined as double alleles in two or more VNTR loci (13, 15). Because mixtures of two strains of the same genotype are virtually impossible to detect by using only RFLP or spoligotyping, we used the VNTR typing results to check the number of potentially missed mixed infections. We defined the occurrence of double alleles at at least two VNTR loci to be potential mixed infections. However, at present, the sensitivity of VNTR typing for the detection of mixed infections is unknown and double alleles could also represent evolution of the bacterium. Therefore, we do not report the mixed-infection rate according to VNTR typing, but we typed all 1,248 isolates by VNTR, RFLP, and spoligotyping to compare their sensitivities for the detection of mixed M. tuberculosis infections as a first step for future studies. Furthermore, VNTR typing was done since the data were also collected for other studies performed to present an overview of genotypes in this part of Vietnam and to check for relapse versus new infections.
Reculture of sputum for analysis of single colonies.
We repeated RFLP and spoligotyping twice for all 39 mixed infections from their DNA (extracted from cultures), and we got the same results. Ideally, to exclude mixed infections that occur due to cross-contamination during the culture processing, studies on mixed infections should be performed on original sputum samples. For two of the assumed mixed-infection isolates, we tried to confirm the observation and exclude laboratory cross-contamination by using a bacteriological approach. Pretreated sputum specimens, stored at −20°C, were recultured on 7H10 agar plates to grow single colonies. Spoligotyping was applied to five or six single colonies of each of the isolates.
Data analysis.
The Gene Marker software, version 1.5 (Softgenetics, PA), was used for analysis and automated allele calling of the VNTR patterns. The Bionumerics software, version 3.0 (Applied Maths, Sint-Martens Latem, Belgium), was used for the analysis and comparison of IS6110 RFLP, spoligotype, and VNTR patterns. Data were entered into Epi Info software, version 6.04 (Centers for Disease Control and Prevention, Atlanta, GA). Double entry was done on a 20% random sample (50% in 2003) of all records. Discrepancies were observed in <1% of all records and in <0.05% of all fields. Analyses were performed in Epi Info, version 6.04, and Stata, version 10SE (Stata Corporation, College Station, TX).
For comparison of categorical variables, we used the chi-square and two-sided Fisher's exact tests as appropriate, with trends across ordered categories assessed by Cuzick's test for trend. Results were considered significant at P values of <0.05. Associations between mixed infections and explanatory variables were expressed as odds ratios; confounding effects were investigated by stratified analysis using the Mantel-Haenszel test.
RESULTS
During the period from January 2003 to December 2005, 1,890 patients were eligible for inclusion, and isolates from 1,248 (66%) of those patients had complete IS6110 RFLP, spoligo-, and VNTR typing results and were available for the analyses. Of these, 931 (74.6%) isolates were from male patients and 317 (25.4%) were from female patients with a median age of 50 years (25th and 75th percentiles, 37 and 66 years, respectively). Eleven hundred seven (88.7%) were new patients, 139 (11.1%) were relapse patients, and the remaining 2 cases were of unknown status.
Identification of mixed infections identified by IS6110 RFLP typing, spoligotyping, and VNTR typing.
Thirty-nine (3.1%) of 1,248 isolates had RFLP and spoligotype patterns that represented different M. tuberculosis genotype families and were considered to be mixed infections. Repeated RFLP and spoligotype analysis from the same DNA confirmed the mixed infections for all 39 isolates. Twenty-eight isolates (71.8%) represented a mixture of Beijing and EAI strains, 5 isolates (12.8%) were a mixture of a Beijing strain and a strain of another genotype (not EAI), 5 isolates (12.8%) were a mixture of an EAI strain and a strain of another genotype (containing more than 4 IS6110 copies in RFLP analysis and not belonging to the Beijing genotype), and 1 (2.6%) was a mixture of a Haarlem and another strain (Fig. 1). Overall, the study population, including the mixed-infection isolates, contained 549 (42.7%) EAI strains, 461 (35.8%) Beijing strains, and 277 (21.5%) strains of other genotypes. These rates of EAI and Beijing genotypes of M. tuberculosis were similar to those reported previously for the prevalence of these genotypes in rural areas of South Vietnam (49% for EAI and 35% for Beijing [5]), but the prevalence of the Beijing genotype was lower than that observed in Hanoi and Ho Chi Minh City (1).
Fig 1.
Mixed infections detected by IS6110 RFLP and spoligotyping. Spoligotypes and IS6110 RFLP patterns of the M. tuberculosis isolates that were identified as mixed infections on the basis of discordant genotyping results. The dendrogram (shown on the left) shows the similarity of the spoligotype patterns, as determined by using the Dice coefficient and unweighted-pair group method using average linkages for clustering. To the right of the IS6110 RFLP patterns, the interpreted genotype families are indicated, as determined by spoligotyping and IS6110 RFLP typing, respectively.
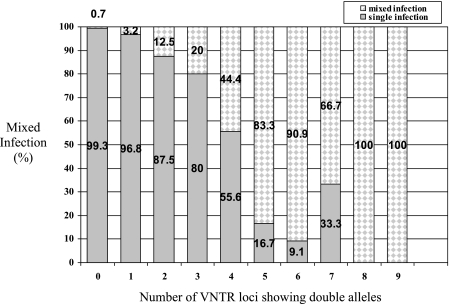
VNTR typing detected 122 (9.8%) of 1,248 isolates with double alleles at at least one locus, and 60 (4.8%) of these isolates revealed double alleles at two or more loci (Table 1). Of these 60 isolates, 31 (51.7%) isolates had not been identified as being mixed infections by combining the results of RFLP and spoligotyping and 29 (48.3%) isolates represented mixed infections confirmed by RFLP and spoligotyping. Thus, of the 39 mixed-infection isolates detected by RFLP and spoligotyping, 29 (74.4%) were confirmed by VNTR typing (Table 1). Comparing the RFLP/spoligotyping and VNTR typing results for the isolates revealed that the percentage of mixed infections detected by RFLP and spoligotyping of isolates strongly increased with the number of loci at which double alleles were found (P < 0.001; Fig. 2). While only 3.2% (2/62) of isolates contained mixed infections (on the basis of RFLP/spoligotyping), when only a single VNTR locus had double alleles, this proportion was more than 80% for isolates that had double alleles in five or more VNTR loci. Among isolates that had two, three, or four loci with double alleles, the proportions of mixed infections were 12.5%, 20.0%, and 44.4%, respectively. Of 1,126 strains having single alleles, 0.7% (8/1,126) were mixed infections, according to combined RFLP and spoligotyping analysis (Table 1). The agreement between the two definitions that we used in this study (discrepant genotype results between RFLP and spoligotyping and double alleles at at least 2 loci with VNTR typing) was 96.7% (Table 1), and Cohen's kappa statistic was 0.57.
Table 1.
Comparison of sensitivity of detection of mixed M. tuberculosis infections by VNTR typing and by combined typing with IS6110 RFLP and spoligotyping
| Pattern by IS6110 RFLP and spoligotyping combined | No. of isolates showing the following by 15-locus VNTR typing: |
|||
|---|---|---|---|---|
| Double alleles at two or more loci | Double allele at one locus | All single alleles | Total | |
| Mixed infection | ||||
| Yes | 29 | 2 | 8 | 39 |
| No | 31 | 60 | 1,118 | 1,209 |
| Total | 60 | 62 | 1,126 | 1,248 |
Fig 2.
Correlation between the heterogeneity in VNTR patterns and the detection of mixed infections by combined IS6110 RFLP and spoligotyping among 1,248 M. tuberculosis isolates. The percentage of mixed infections by (IS6110 RFLP/spoligotyping) is indicated for isolates showing VNTR patterns with no double alleles (n = 1,126), a double allele at 1 VNTR locus (n = 62), or double alleles at 2 to 9 different VNTR loci (n = 60).
Of the 60 isolates that had two or more double alleles in VNTR typing, 29 represented a mixed infection on the basis of the combination of the RFLP and spoligotype results; 25 of these were a mixture of Beijing and EAI genotypes, 3 were a mixture of EAI and another genotype, and 1 was a mixture of a Beijing and another genotype. For 31 of the 60 isolates that had two or more double alleles in VNTR typing, no mixed infection was detected with RFLP and spoligotyping.
Reculturing of mixed-infection isolates.
Since identification of mixed infection by culture was not the primary purpose of this study, no attention was given to the phenotypic nature of the cultures. Only after we discovered several samples to contain mixed infections did we decide to reculture these. Unfortunately, enough material to reculture the sputum sample was available for only two samples. For two of the assumed mixed infections for which original sputum specimens were still available, pretreated sputum specimens were recultured directly on 7H10 agar plates and spoligotyping was applied to five or six single colonies of each of these two isolates. For one specimen, three colonies yielded Beijing-specific spoligotype patterns, while two other colonies revealed an EAI spoligotype. For the other specimen, only Beijing spoligotypes were obtained. Thus, a mixed infection was confirmed microbiologically for one of the two isolates.
In terms of microbiology, we observed that the colonies of Beijing genotype isolates were often smooth and large, with a diameter of more than 2 mm, while the colonies of EAI genotype strains were often dry and small, with a diameter of less 1 mm.
Epidemiological and clinical characteristics of mixed infections.
There were no significant associations between the probability of having a mixed infection and the district of residence, age, sex, treatment history, treatment delay, presence of systemic symptoms, treatment outcome, or the isolates' resistance to isoniazid or streptomycin or multidrug resistance (Table 2). However, mixed infections were significantly less likely to occur in patients with extensive X-ray abnormalities (P < 0.001), and there was a nearly significant trend for lower sputum smear grades among mixed infections (P = 0.054), with both findings suggesting less extensive pathology in patients with mixed infections. Taking a medium severity of X-ray abnormalities as the reference, the odds of a mixed infection was 2.52 times (95% CI, 1.08 to 5.87) higher for minor X-ray abnormalities and 0.42 times (95% CI, 0.19 to 0.92) lower for major X-ray abnormalities. For sputum smear grading, taking a 1+ grade as the baseline, the odds of mixed infection was not higher for negative or scanty smears (1.10 times; 95% CI, 0.48 to 2.52) but was 0.48 time lower for 2+ or 3+ smears (95% CI, 0.22 to 1.07). In stratified analyses, the odds ratios for the association of mixed infection with either severity of X-ray abnormalities or sputum smear grade were not affected by any of the variables in Table 2, nor were X-ray abnormalities or sputum smear grade clearly associated with a specific genotype (EIA, Beijing, or other) among the single infections (data not shown).
Table 2.
Patient and TB disease characteristics for patients with mixed M. tuberculosis infections
| Characteristic | No. (%) of patients |
P valuea | |
|---|---|---|---|
| Total | Mixed infections | ||
| Total | 1,248 | 39 (3.1) | |
| District | |||
| Cai Lay | 575 | 22 (3.8) | 0.299 |
| Chau Thanh | 392 | 8 (2.0) | |
| Cai Be | 281 | 9 (3.2) | |
| Sex | |||
| Male | 931 | 29 (3.1) | 1.000 |
| Female | 317 | 10 (3.2) | |
| Age (yr) | |||
| 15-34 | 246 | 9 (3.7) | 0.884 |
| 35-64 | 657 | 20 (3.0) | |
| ≥65 | 344 | 10 (2.9) | |
| Unknown | 1 | 0 (0) | |
| Treatment history | |||
| New | 1,107 | 38 (3.4) | 0.17 |
| Previously treated | 139 | 1 (0.7) | |
| Unknown | 2 | 0 (0) | |
| Treatment outcome | |||
| Favorable | 1,226 | 39 (3.2) | 1.000 |
| Unfavorable | 23 | 0 (0) | |
| Abnormalities on X ray | |||
| Minor | 90 | 8 (8.9) | 0.001 |
| Medium | 590 | 22 (3.7) | |
| Major | 562 | 9 (1.6) | |
| Unknown | 6 | 0 (0) | |
| Smear gradeb | |||
| Negative or scanty | 194 | 8 (4.1) | 0.117c |
| 1+ | 557 | 21 (3.8) | |
| 2-3+ | 484 | 9 (1.9) | |
| Missing | 13 | 1 (7.7) | |
| Duration of cough (wk) | |||
| <4 | 324 | 10 (3.1) | 0.978 |
| 4-7 | 469 | 14 (3.0) | |
| ≥8 | 455 | 15 (3.3) | |
| Fever | |||
| Present | 1,108 | 36 (3.2) | 0.613 |
| Absent | 140 | 3 (2.1) | |
| Night sweats | |||
| Present | 748 | 27 (3.6) | 0.249 |
| Absent | 500 | 12 (2.4) | |
| Weight loss | |||
| Present | 1,223 | 38 (3.1) | 0.551 |
| Absent | 25 | 1 (4.0) | |
| Isoniazid susceptibility | |||
| Resistant | 246 | 8 (3.3) | 0.840 |
| Sensitive | 1,002 | 31 (3.1) | |
| Streptomycin susceptibility | |||
| Resistant | 341 | 15 (4.4) | 0.142 |
| Sensitive | 907 | 24 (2.6) | |
| Multidrug resistant | |||
| Yes | 50 | 3 (6.0) | 0.203 |
| No | 1,198 | 36 (3) | |
Calculated by Fisher's exact test.
The results in the reference lab; maximum grading of two smear examinations.
Cuzick's test for trend, P = 0.054.
On the basis of demographic information, epidemiological links between patients with mixed infections were considered highly unlikely. Patients with mixed infections were distributed equally over the three different provinces; lived in different communities; were from different sexes, age classes, and socioeconomic classes; had different types of jobs; and did not occur within the same household (data not shown).
DISCUSSION
Our study shows that mixed tuberculosis infections also occur outside settings with extremely high TB incidences and population densities. The proportion of mixed tuberculosis infections found in rural South Vietnam in this study was 3.1% when identified by combined RFLP and spoligotyping results and 4.8% when identified by ≥2 loci with double alleles in VNTR typing, which are similar to the proportion reported from South Africa by Richardson et al. (2.3%) (14) but lower than that found in Georgia (13.1%) (15) or in South Africa by Warren et al. (19%) (22) and Cohen et al. (9%) (7). These differences in the proportions of mixed infections could be explained by differences in the methodology of detection. The study from Georgia defined mixed infections on the basis of RFLP typing as well as VNTR typing (15). If we would have used a definition for mixed infections adding RFLP/spoligotyping and VNTR results, our study would have identified (39 + 31)/1,248, or 5.6%, mixed infections. Moreover, a number of mixed infections in the Georgian study were detected only after typing of multiple specimens from a single patient (15), and the study by Cohen et al. in South Africa focused on pooled multiple samples from autopsy patients (7), while we typed only one specimen per patient. The study in South Africa by Warren et al. (22) used a methodology based on PCR probes distinguishing Beijing from non-Beijing strains which is more sensitive for identifying mixed infections than the RFLP typing-based method applied by Richardson et al. (14) that we also used. In addition, differences in the densities of human populations and risk of TB infection are likely to have played a role. The study in Georgia took place in a crowded prison with a TB incidence of 5,995/100,000, the study by Warren et al. was performed in an urban area with a TB incidence of as high as 1,000/100,000 (22), and the study by Cohen et al. was performed in a highly HIV-infected hospital population (7). In these settings, cross-infection could easily occur. In contrast, our study was performed in a rural area in Vietnam with a population density of only 837/km2 and an observed TB incidence of new smear-positive cases of 100/100,000 in 2005 (National Tuberculosis Program Vietnam, unpublished data), and cross-infection may be expected to be less common under these circumstances. Finally, HIV infection may increase the potential for acquiring mixed TB infections (16), and the prevalence of HIV infection among TB patients was higher in the South African studies (at least 10% [22] and 94% [7]) than in ours.
To identify mixed infections in our study, we used RFLP and spoligotyping, in which strains of the EAI and Beijing genotypes can be recognized easily. It is likely that mixed infections between a Beijing and an EAI strain are easier to detect by a Beijing RFLP pattern and an EAI spoligotype than the other way around because the Beijing RFLP pattern can mask the EAI RFLP pattern and the EAI spoligotype can mask the Beijing spoligotype. Therefore, the combination of methods is necessary to detect the mixture. The Beijing RFLP type and EAI spoligotype mixture is also the mixture that we most frequently detected in this study. We did not find any mixtures of the reverse type: an EAI RFLP type and a Beijing spoligotype. This is because an EAI strain can be visualized by RFLP typing only when the Beijing strain is present in a very low concentration compared to the EAI strain in a sample. In such a case, in spoligotyping, the EAI strain will also be amplified and visible in the spoligotyping pattern. The spoligotype of the Beijing strain that would be present in the sample would be masked by the EAI spoligotype. RFLP and VNTR typing have been used before by Shamputa et al. to detect mixed infections (15). Our study showed that RFLP and spoligotyping can also be used for detection of mixed infections.
A disadvantage of our approach is that we probably underestimated the true rate of mixed infections in Vietnam. The fact that we detected 60 strains with double alleles in at least two VNTR loci suggests that VNTR typing is a more sensitive method to detect mixed infections than RFLP/spoligotyping. Neither RFLP nor spoligotyping can independently detect a mixture of two strains of the same genotype very efficiently (8), whereas VNTR typing results can show double alleles at chromosomal loci and thus in principle also detect multiple strains of the same genotype. On the other hand, 8/39 (21%) of the mixed infections were missed by VNTR typing (Table 1). We found that the more loci with double alleles an isolate showed in VNTR typing, the higher the probability was that these were also identified as mixtures by combined RFLP and spoligotyping (Fig. 2). At present, the sensitivity of VNTR typing for the detection of mixed infections is unknown, and double alleles could also represent evolution of the bacterium (13); therefore, we did not define the mixed-infection rate according to VNTR results, but we typed all 1,248 isolates by VNTR, RFLP, and spoligotyping to compare the sensitivities of these methods to detect mixed M. tuberculosis infections as a first step for future studies.
With 100% of the cases being cured, we do not have an indication that mixed infections led to higher failure rates. In fact, they were associated with less extensive pulmonary pathology than single infections. Although this is based on routine chest X-ray results that were not standardized as part of our study, we found this association across all three districts, suggesting that it does not reflect observer bias. Moreover, we found a similar pattern with regard to sputum smear grading (which had been done centrally): mixed infections were associated with a lower degree of smear positivity. Interestingly, neither of these associations was determined by underlying differences in duration of cough or presence of systemic symptoms such as fever, night sweats, and weight loss. This suggests that the observed associations do not reflect early diagnosis (i.e., the patient still had limited pulmonary pathology), for example, because of a higher incidence of systemic symptoms with mixed than with single infections. This finding requires further studies; one hypothesis could be that patients with multiple infections have an increased immunological tolerance to M. tuberculosis infections. This may be due to HIV infection, for which we did not test. However, other causes are likely to play a role as well, since mixed infections occurred equally among both sexes and all age groups, while HIV infection among TB patients in Vietnam is strongly associated with young age (<35 years) and male sex (18). It is also interesting that mixed-infection cases were not related to a history of TB treatment, as they occurred in 3.4% of the new patients, compared to 0.72% of the patients with recurrent TB. This finding was in agreement with the report from Georgia (15). In contrast, Warren et al. (22) found that multiple infections were more frequent in relapse cases.
Although we cannot completely exclude the possibility that (part of) the mixed infections detected in our study were the result of errors or cross-contamination in the laboratory, various observations support our finding. The mixed infections were found in all three districts, with the highest frequency of mixed infections being in the district with the highest TB rate, and at different points in time. Furthermore, repeated analysis with all three DNA typing methods invariably confirmed the initially obtained results. Finally, to check for the possibility of cross-contamination, we recultured single colonies of the only two sputum specimens that were still available and could confirm mixed bacterial populations in one of these two with typing of only a very limited number of single colonies. This suggests that the observed discordant results between RFLP and spoligotyping results indeed represented mixed infections.
There were some limitations to our study. First, we did not perform HIV testing, so we could not study the relationship between HIV and mixed infections, although at 0.5% the HIV prevalence is still very low in rural Vietnam (19). Second, we did not store all sputum samples for reculture to recheck for mixed infections. Third, the ability of RFLP and spoligotyping to detect mixtures of strains with the same genotype is limited. Fourth, VNTR typing is thought to be more sensitive than RFLP and spoligotyping to detect mixed infections, but the exact sensitivity of VNTR typing for the detection of mixed infections is unknown. Finally, the number of mixed infections that we identified was relatively small, thereby limiting the power of our study to detect significant associations with potential risk factors.
We believe that the observed 3.1% of mixed infections in the rural part of South Vietnam represents a minimum estimate, as the approach explored by combining RFLP and spoligotyping has a restricted detection limit. Most likely, due to the visual aspect involved in this approach, mixed infections that have more uneven ratios between the number of bacteria of the respective strains will go unnoticed. With the application of VNTR typing, which may be more sensitive in the detection of mixed infection, the true magnitude of this phenomenon may be unraveled in the future.
ACKNOWLEDGMENTS
This research was funded by KNCV Tuberculosis Foundation, the Medical Committee Netherlands-Vietnam, The Netherlands, and supported by the TBadapt project (EC 6th Framework project number 037919).
We thank all TB patients participating in this study and the staff of the National Tuberculosis Program of Tien Giang Province for recruiting the patients as well as the staff of the National Tuberculosis Program of Pham Ngoc Thach Tuberculosis and Lung Disease Hospital for supervision and checking of the data. Dai Viet Hoa, Phan Thi Hoang Anh, and other staff in the Pham Ngoc Thach laboratory are thanked for performing culture, spoligotyping, RFLP, and VNTR typing. We are grateful to Anne-Marie van den Brandt, Mimount Enaimi, and Arnout Mulder and other staff of the Tuberculosis Reference Laboratory at the National Institute for Public Health and the Environment (The Netherlands) for providing us with the knowledge of and practice with the molecular techniques used in this study. We thank the staff at the laboratory of the Oxford University Clinical Research Unit (Ho Chi Minh City) for their help in performing the VNTR typing.
This study was performed at the Pham Ngoc Thach Tuberculosis and Lung Disease Hospital, Ho Chi Minh City, Vietnam.
Footnotes
Published ahead of print 29 February 2012
REFERENCES
- 1. Anh DD, et al. 2000. Mycobacterium tuberculosis Beijing genotype emerging in Vietnam. Emerg. Infect. Dis. 6:302–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bates JH, Stead WW, Rado TA. 1976. Phage type of tubercle bacilli isolated from patients with two or more sites of organ involvement. Am. Rev. Respir. Dis. 114:353–358 [DOI] [PubMed] [Google Scholar]
- 3. Braden CR, et al. 2001. Simultaneous infection with multiple strains of Mycobacterium tuberculosis. Clin. Infect. Dis. 33:e42–e47 [DOI] [PubMed] [Google Scholar]
- 4. Brudey K, et al. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buu TN, et al. 2009. The Beijing genotype is associated with young age and multidrug-resistant tuberculosis in rural Vietnam. Int. J. Tuberc. Lung Dis. 13:900–906 [PubMed] [Google Scholar]
- 6. Buu TN, et al. 2010. Tuberculosis acquired outside of households, rural Vietnam. Emerg. Infect. Dis. 16:1466–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cohen T, Wilson D, Wallengren K, Samuel EY, Murray M. 2011. Mixed-strain M. tuberculosis infections among patients dying in hospital in KwaZulu-Natal, South Africa. J. Clin. Microbiol. 49:385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Boer AS, et al. 2000. Genetic heterogeneity in Mycobacterium tuberculosis isolates reflected in IS6110 restriction fragment length polymorphism patterns as low-intensity bands. J. Clin. Microbiol. 38:4478–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoa NB, et al. 2010. National survey of tuberculosis prevalence in Viet Nam. Bull. World Health Organ. 88:273–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. International Union against Tuberculosis and Lung Disease 2000. Technical guide-sputum examination for tuberculosis direct microscopy in low-income countries. International Union against Tuberculosis and Lung Disease, Paris, France [Google Scholar]
- 11. Kamerbeek J, et al. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kremer K, et al. 2004. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J. Clin. Microbiol. 42:4040–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martin A, Herranz M, Serrano MJ, Bouza E, Garcia de Viedma D. 2007. Rapid clonal analysis of recurrent tuberculosis by direct MIRU-VNTR typing on stored isolates. BMC Microbiol. 7:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Richardson M, et al. 2002. Multiple Mycobacterium tuberculosis strains in early cultures from patients in a high-incidence community setting. J. Clin. Microbiol. 40:2750–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shamputa IC, et al. 2006. Mixed infection and clonal representativeness of a single sputum sample in tuberculosis patients from a penitentiary hospital in Georgia. Respir. Res. 7:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Small PM, et al. 1993. Exogenous reinfection with multidrug-resistant Mycobacterium tuberculosis in patients with advanced HIV infection. N. Engl. J. Med. 328:1137–1144 [DOI] [PubMed] [Google Scholar]
- 17. Supply P, et al. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thanh DH, et al. 2010. HIV infection among tuberculosis patients in Vietnam: prevalence and impact on tuberculosis notification rates. Int. J. Tuberc. Lung Dis. 14:986–993 [PubMed] [Google Scholar]
- 19. Thuy TT, et al. 2007. HIV-associated TB in An Giang Province, Vietnam, 2001-2004: epidemiology and TB treatment outcomes. PLoS One 2:e507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Embden JD, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Soolingen D, de Haas PEW, Kremer K. 2001. Restriction fragment length polymorphism typing of mycobacteria, p 165–203 In Parish T, Stoker NG. (ed), Mycobacterium tuberculosis protocols. Humana Press Inc., Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 22. Warren RM, et al. 2004. Patients with active tuberculosis often have different strains in the same sputum specimen. Am. J. Respir. Crit. Care Med. 169:610–614 [DOI] [PubMed] [Google Scholar]
- 23. World Health Organization 2003. Guidelines for surveillance of drug resistance in tuberculosis. World Health Organization, Geneva, Switzerland [Google Scholar]
- 24. World Health Organization 2003. Treatment of tuberculosis. Guidelines for national programmes. World Health Organization, Geneva, Switzerland [Google Scholar]
- 25. Yeh RW, Hopewell PC, Daley CL. 1999. Simultaneous infection with two strains of Mycobacterium tuberculosis identified by restriction fragment length polymorphism analysis. Int. J. Tuberc. Lung Dis. 3:537–539 [PubMed] [Google Scholar]