Abstract
A real-time multiplex PCR targeting stx2, wzyO104, and fliCH4 of enterohemorrhagic Escherichia coli (EHEC) O104:H4 correctly determined the presence or absence of these genes in 253 EHEC isolates and enrichment cultures of stool samples from 132 patients. It is a rapid, sensitive, and specific tool for detecting EHEC O104:H4 in human stools.
TEXT
A large outbreak caused by Shiga toxin 2 (Stx2)-producing enterohemorrhagic Escherichia coli (EHEC) O104:H4 that involved >4,000 cases, including 855 patients with hemolytic-uremic syndrome (HUS) and 53 fatalities, occurred in Germany in May to July 2011 (6, 14). Early in the outbreak, we and others developed PCRs targeting typical molecular features of the outbreak strain, including the stx2 gene and the wzyO104 and fliCH4 genes, components of the O104 and H4 antigen synthetic gene clusters, respectively (3), or stx2 plus aggC and aggD, components of the gene cluster encoding aggregative adherence fimbria I (13). Here, we extended this approach by developing a real-time multiplex PCR (rtMPCR) and evaluating its utility for the detection of EHEC O104:H4 in human stools.
The rtMPCR was performed in a CFX96 real-time PCR system (Bio-Rad, Munich, Germany) in a 20-μl volume containing 10 μl of 2× Fast EvaGreen SuperMix (Bio-Rad), each primer in a concentration optimized (Table 1) to obtain melting peaks of similar intensities, and 1 μl (∼30 ng) of genomic DNA extracted using InstaGene Matrix (Bio-Rad). The cycling protocol included initial denaturation at 98°C for 2 min, as recommended by the manufacturer, followed by 35 cycles of denaturation (98°C, 10 s), annealing (55°C, 10 s), and extension (72°C, 20 s). Fluorescence was recorded at the end of each extension. Melting curves were generated from 65°C to 95°C with increments of 0.2°C/s. Results were analyzed using Bio-Rad CFX Manager version 1.6. Positive (EHEC O104:H4), negative (E. coli K-12 C600), and no-template (distilled water) controls were included in each run.
Table 1.
Primers for real-time multiplex PCR to identify stx2-harboring E. coli O104:H4
| Primera | Sequence (5′–3′) | Concn (nM) per reactionb | Target | Amplicon size (bp) |
|---|---|---|---|---|
| O104wzy-f | GGTTATGTTCTTGTCTTTGC | |||
| O104wzy-r | CTAATACTTGTCTGATACGG | 225 | wzyO104 | 154 |
| RT-stx2F | CGACCCCTCTTGAACATA | |||
| RT-stx2R | TAGACATCAAGCCCTCGTAT | 100 | stx2 | 106 |
| fliCH4-a | GGCGAAACTGACGGCTGCTG | |||
| fliCH4-b | GCACCAACAGTTACCGCCGC | 75 | fliCH4 | 201 |
Primers for wzyO104 and stx2 were designed in this study. Primers for fliCH4 were described previously (3).
The primer concentrations were optimized to obtain melting peaks of similar intensities.
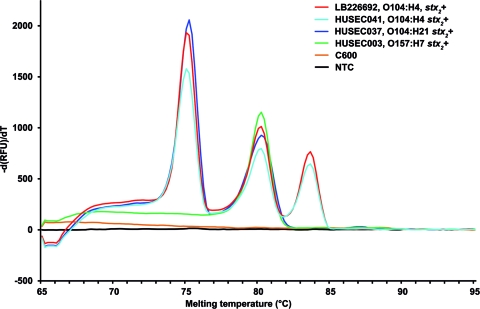
A total of 253 EHEC isolates, including 91 O104:H4 outbreak isolates, 42 strains of the HUSEC (HUS-associated E. coli) collection (10) (www.ehec.org), which includes prototypic strains of all EHEC serotypes associated with HUS in Germany (see Table S1 in the supplemental material), and 120 EHEC strains that represent the whole spectrum of diarrhea-associated EHEC serotypes present in our collections (see Table S2 in the supplemental material) were tested to determine the specificity of the rtMPCR. Serotyping, stx typing, and multilocus sequence typing (MLST) of the strains were performed as described previously (2, 4, 10, 12). All 91 EHEC O104:H4 outbreak isolates produced all three amplicons, with melting peaks at 75.2°C for wzyO104, 80.2°C for stx2, and 83.6°C for fliCH4 (Fig. 1). In the HUSEC collection, these three peaks were produced only by strain HUSEC041 (stx2-harboring EHEC O104:H4) (Fig. 1; see also Table S1 in the supplemental material). Another strain, HUSEC037 (stx2-harboring EHEC O104:H21), yielded amplicons for wzyO104 and stx2 but not for fliCH4 (Fig. 1; see also Table S1 in the supplemental material). In addition, strain HUSEC038, which belongs to serotype Ont (nontypeable O antigen):H21 and to the same multilocus sequence type (ST672) as HUSEC037, also produced the wzyO104 but not the fliCH4 amplicon (see Table S1 in the supplemental material). The specificity of the wzyO104 amplification in HUSEC038 was confirmed by partial (627-bp) sequencing of the gnd locus, which is adjacent to the O104 antigen-encoding cluster and is serogroup specific (7). The partial gnd sequences of HUSEC038 and HUSEC037 were 100% identical to that of HUSEC041, demonstrating that HUSEC038 indeed belongs to serogroup O104. Among the 120 EHEC strains associated with diarrhea, the rtMPCR amplified wzyO104 in the strain of serotype O104:H21 and fliCH4 in all strains with the H4 antigen (serotypes O8:H4, O68:H4, O113:H4, and O119:H4) as well as in two nonmotile strains (O74:H− and O78:H−), each of which contained fliCH4, as demonstrated by fliC restriction fragment length polymorphism analysis (19) (see Table S2 in the supplemental material). In all HUSEC strains and the diarrhea-associated EHEC strains, the stx2 primer pair detected stx2 and its variants stx2c, stx2d, stx2dactivatable, and stx2e but not stx2f (see Tables S1 and S2 in the supplemental material), which has a low degree of sequence homology to stx2 (15); however, that allele is only rarely present in human clinical EHEC (16).
Fig 1.
Real-time multiplex PCR for the detection and identification of EHEC O104:H4. Data represent amplification of wzyO104, stx2, and fliCH4 in prototypic EHEC O104:H4 outbreak isolate LB226692 (3, 11), HUSEC041 (O104:H4), HUSEC037 (O104:H21), and HUSEC003 (O157:H7) by the use of the real-time MPCR. Melting peaks of the wzyO104, stx2, and fliCH4 amplicons at 75.2°C, 80.2°C, and 83.6°C, respectively, are shown. C600, E. coli K-12 C600; NTC, no-template control.
To determine the utility of the rtMPCR for the detection of EHEC O104:H4 in human stools, we applied the test to 132 stool samples from patients with HUS or diarrhea, which were selected from 1,207 stool samples received in our laboratory between 23 May and 31 July 2011; 412 of these stools were positive for the outbreak strain, as determined by culture. For the rtMPCR, every 5th culture-positive sample and every 15th culture-negative stool sample were used. To prepare PCR templates, the stool samples were enriched for 4 h in GN broth Hajna (Difco Laboratories, Detroit, MI) and a 100-μl volume was plated on individual plates of sorbitol MacConkey agar (Becton Dickinson, Heidelberg, Germany), enterohemolysin agar (Sifin, Berlin, Germany), and extended-spectrum β-lactamase agar (chromID ESBL; bioMérieux, Nurtingen, Germany). The overnight growth from the plates was washed into 1 ml of 0.9% NaCl solution and boiled for 10 min; 1 μl of the total extracted DNA (diluted 1:10 in sterile water) was used per 20 μl of rtMPCR volume. Eighty-three (62.9%) of the 132 stool enrichment cultures contained the EHEC O104:H4 outbreak strain, as demonstrated by conventional multiplex PCR (3) and subsequent isolation of the strain, which was used as the gold standard to which results of the rtMPCR were compared. Each of the 83 cultures produced all three amplicons (wzyO104, stx2, and fliCH4) in the rtMPCR. The remaining 49 enrichment cultures lacked the outbreak strain in both the conventional multiplex PCR and culture on ESBL agar. Thirty-eight of them yielded none of the three amplicons, whereas 11 yielded the stx2 amplicon only; various stx2-positive non-O104:H4 E. coli strains were subsequently isolated from these 11 samples. The detection limit of the rtMPCR for identification of EHEC O104:H4 in stool cultures was determined by spiking three different human O104:H4-negative stools enriched for 4 h in GN broth Hajna with 10-fold dilutions (101 to 1010 CFU/ml) of EHEC O104:H4 outbreak strain LB226692 (3, 11), growing 100 μl of the mixtures on ESBL agar and Luria-Bertani agar plates at 37°C overnight, extracting total DNA from bacteria washed from the plates by boiling for 10 min, and using 1 μl of the DNA in rtMPCR. The detection limit of the rtMPCR was 7 × 103 (range, 1 × 103 to 1 × 104) CFU/ml of EHEC O104:H4 on the background of 4.2 × 107 (range, 7 × 106 to 6 × 107) CFU/ml of normal coliform intestinal flora. The detection limit of the test for EHEC O104:H4 strain LB226692 in pure culture was 1.6 × 102 CFU/ml.
The rtMPCR developed here has 100% specificity and 100% sensitivity for the detection of EHEC O104:H4 in human stool samples compared to culture (i.e., isolation of the strain) and for identification of EHEC O104:H4 isolates compared to serotyping. Although the EHEC O104:H4 outbreak is over, this assay can be utilized in diagnostic laboratories in Germany, in particular, in those specialized for detection of EHEC, because sporadic cases of infection with the outbreak strain still rarely occur in this country (our unpublished data). Also, the rtMPCR represents a rapid and reliable tool for epidemiological studies to determine the prevalence of EHEC O104:H4 in the human population, which is considered the major (if not the only) reservoir of this pathogen (1). Moreover, because the rtMPCR detects stx2 and its variants present in HUS-associated as well as diarrhea-associated EHEC (see Tables S1 and S2 in the supplemental material), it will also detect non-O104:H4 EHEC causing human disease. Thus, stool samples positive only for stx2 in the rtMPCR, as was the case for the samples from the 11 patients described above, need to be further investigated for EHEC of other serotypes in order to detect both known and possibly new, emerging EHEC strains. Preliminary information about the presence of an EHEC strain in the stool within 24 h, as provided by the rtMPCR, is critical for epidemiological purposes, in particular, for “real-time” monitoring of spread of the infection and tracing it back to the source. From the therapeutic standpoint, rapid detection of evidence of EHEC O104:H4 infection may provide a basis for applying, in addition to general therapeutic strategies used for EHEC infections (increasing the volume of intravenous fluids and avoiding antibiotic administration) (9, 17), additional, more specialized approaches such as were successfully used during the EHEC O104:H4 outbreak (8). The method described here extends the real-time multiplex PCRs available for detecting EHEC O104:H4 in food (5, 18) for the first time to rapid detection of the strain in human stools. rtMPCRs for detecting other members of the HUSEC collection are under development.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from the Interdisciplinary Center of Clinical Research (IZKF) Münster (Me2/021/12) and the Medical Faculty of the University of Münster (BD9817044).
We thank Ralph Fischer and Andrea Lagemann for technical assistance.
Footnotes
Published ahead of print 15 February 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Beutin L, Martin A. 2012. Outbreak of Shiga toxin-producing Escherichia coli (STEC) O104:H4 infection in Germany causes a paradigm shift with regard to human pathogenicity of STEC strains. J. Food Prot. 75:408–418 [DOI] [PubMed] [Google Scholar]
- 2. Bielaszewska M, Friedrich AW, Aldick T, Schürk-Bulgrin R, Karch H. 2006. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin. Infect. Dis. 43:1160–1167 [DOI] [PubMed] [Google Scholar]
- 3. Bielaszewska M, et al. 2011. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 11:671–676 [DOI] [PubMed] [Google Scholar]
- 4. Bielaszewska M, et al. 2009. Shiga toxin, cytolethal distending toxin, and hemolysin repertoires in clinical Escherichia coli O91 isolates. J. Clin. Microbiol. 47:2061–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. EUReference Laboratory for Ecoli (EU-RL VTEC) 2 June 2011, posting date Detection and identification of verocytotoxin-producing Escherichia coli (VTEC) O104:H4 in food by real time PCR. Istituto Superiose di Sanità, Rome, Italy: http://www.iss.it/binary/vtec/cont/Lab_Proc_VTEC_O104.pdf [Google Scholar]
- 6. Frank C, et al. 2011. Epidemic profile of Shiga toxin-producing Escherichia coli O104:H4 outbreak in Germany. N. Engl. J. Med. 19:1771–1780 [DOI] [PubMed] [Google Scholar]
- 7. Gilmour MW, Olson AB, Andrysiak AK, Ng L, Chui L. 2007. Sequence-based typing of genetic targets encoded outside of the O-antigen gene cluster is indicative of Shiga toxin-producing Escherichia coli serogroup lineages. J. Med. Microbiol. 56:620–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Greinacher A, et al. 2011. Treatment of severe neurological deficits with IgG depletion through immunoadsorption in patients with Escherichia coli O104:H4-associated haemolytic uraemic syndrome: a prospective trial. Lancet 378:1166–1173 [DOI] [PubMed] [Google Scholar]
- 9. Hickey CA, et al. 2011. Early volume expansion during diarrhea and relative nephroprotection during subsequent hemolytic uremic syndrome. Arch. Pediatr. Adolesc. Med. 165:884–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mellmann A, et al. 2008. Analysis of collection of hemolytic uremic syndrome-associated enterohemorrhagic Escherichia coli. Emerg. Infect. Dis. 14:1287–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mellmann A, et al. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prager R, Strutz U, Fruth A, Tschäpe H. 2003. Subtyping of pathogenic Escherichia coli strains using flagellar H-antigens: serotyping versus fliC polymorphisms. Int. J. Med. Microbiol. 292:477–486 [DOI] [PubMed] [Google Scholar]
- 13. Qin J, et al. 2011. Identification of the Shiga toxin-producing Escherichia coli O104:H4 strain responsible for a food poisoning outbreak in Germany by PCR. J. Clin. Microbiol. 49:3439–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Robert Koch Institute 2011. Report: final presentation and evaluation of epidemiological findings in the EHEC O104:H4 outbreak, Germany 2011. Robert Koch Institute, Wernigerode, Germany: http://www.rki.de/cln_226/nn_217400/EN/Home/EHEC__final__report,templateId=raw,property=publicationFile.pdf/EHEC_final_report.pdf [Google Scholar]
- 15. Schmidt H, et al. 2000. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 66:1205–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sonntag AK, Zenner E, Karch H, Bielaszewska M. 2005. Pigeons as a possible reservoir of Shiga toxin 2f-producing Escherichia coli pathogenic to humans. Berl. Munch. Tierarztl. Wochenschr. 118:464–470 [PubMed] [Google Scholar]
- 17. Tarr PI, Gordon CA, Chandler WL. 2005. Shiga toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365:1073–1086 [DOI] [PubMed] [Google Scholar]
- 18. Tzschoppe M, Martin A, Beutin L. 2012. A rapid procedure for the detection and isolation of enterohaemorrhagic Escherichia coli (EHEC) serogroup O26, O103, O111, O118, O121, O145 and O157 strains and the aggregative EHEC O104:H4 strain from ready-to-eat vegetables. Int. J. Food Microbiol. 152:19–30 [DOI] [PubMed] [Google Scholar]
- 19. Zhang W, et al. 2007. Structural and functional differences between disease-associated genes of enterohaemorrhagic Escherichia coli O111. Int. J. Med. Microbiol. 297:17–26 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.