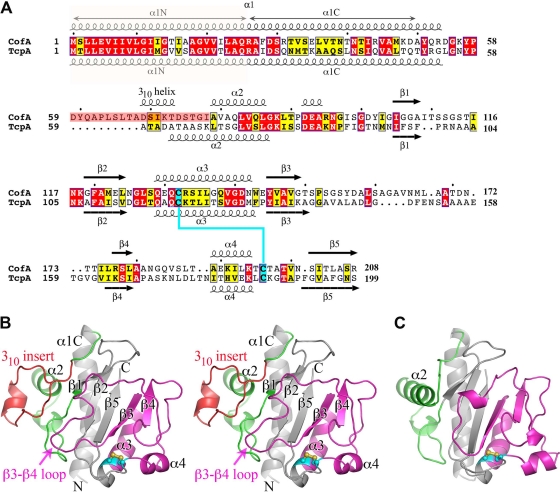
Fig 2.
Comparison of amino acid sequences and atomic structures of ETEC CofA and V. cholerae TcpA. (A) Amino acid sequence alignment of ETEC CofA (strain 31-10; matches GenBank accession number BAA07174) and V. cholerae TcpA (classical strain O395; GenBank accession number ABQ19609), based on alignment of the two crystal structures. Only the mature regions of the pilin proteins are shown. The secondary structure is indicated above and below the sequences. The α1N α-helix was predicted based on amino acid sequence homology with the corresponding regions in the full-length pilin crystal structures (PDB ID 1OQW, 2HI2). Identical residues are shown in white text with red shading; homologous residues are shown in black text with yellow shading. The 310 insert is shaded pink in CofA. Disulfide-bonded cysteines are shaded cyan. (B) Crystal structure of ΔN-CofA (residues 28 to 208) at 1.26-Å resolution, shown in stereo view with the αβ-loop in green and the D-region in magenta. The 310 insert within the αβ-loop is shown in red. Side chains for cysteines Cys132 and Cys196 are shown in a ball-and-stick representation. (C) ΔN-TcpA crystal structure (PDB ID 1OQV), shown for comparison with ΔN-CofA, colored as described for panel B. Side chains for cysteines Cys120 and Cys186 are shown in a ball-and-stick representation.