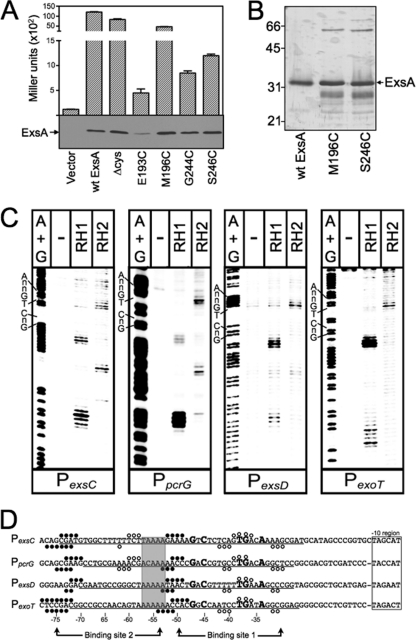
Fig 2.
Fe-BABE footprints of the ExsA RH1- and RH2-Fe-BABE conjugants bound to the PexsC, PpcrG, PexsD, and PexoT promoter probes. (A) The PA103 exsA::Ω strain carrying a PexsD-lacZ reporter fusion was transformed with either a vector control or plasmids expressing wt ExsA, ExsA lacking all seven of the native cysteine residues (Δcys), or ExsA Δcys with unique cysteines introduced at positions E193, M196, G244, or S246. Transformants were grown under inducing conditions for T3SS gene expression and assayed for β-galactosidase activity. Whole-cell lysates (normalized by cell number) were immunoblotted with ExsA antiserum to examine steady-state expression levels. (B) Silver-stained SDS-polyacrylamide gel of wt ExsAHis and the ExsAHis M196C and ExsAHis S246C mutants following purification by Ni2+-affinity chromatography (1 μg each). Note that wt ExsA was further purified using a heparin column. The migration points of molecular weight standards are indicated on the left side of the gel. (C) The indicated promoter probes labeled with 32P on the forward strand were incubated in the absence (−) or presence of the RH1- or RH2-Fe-BABE conjugants for 15 min at 25°C as indicated. Binding reactions were subjected to Fe-BABE cleavage, separated by electrophoresis, and visualized by phosphorimaging. Maxam-Gilbert sequencing ladders (A+G) were included for orientation, and the positions of the GnC and TGnnA sequences in ExsA binding site 1 are indicated. (D) Summary of the Fe-BABE footprinting data for the forward (Fig. 2C) and reverse (data not shown) strands. The −10 hexamers (boxed), conserved GnC and TGnnA sequences (bold with larger typeface), adenine-rich region (gray box), and position of ExsA binding sites 1 and 2 are indicated. The regions of PexsC, PexsD, and PexoT previously shown to be protected from DNase I cleavage by ExsA are underlined (6). The protected region of PpcrG is based upon the DNase I footprint presented in Fig. S1 in the supplemental material. The cleavage events generated by the RH1- and RH2-Fe-BABE conjugants are indicated with filled and open circles, respectively. The open and filled circles located above and below the nucleotide sequence represent the Fe-BABE footprints for the forward and reverse strands, respectively.