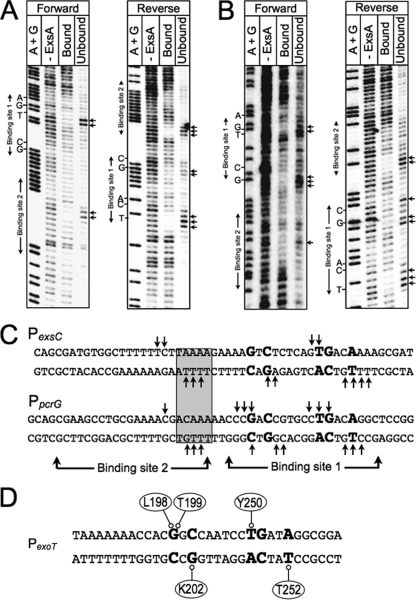
Fig 9.
Missing-nucleoside footprints of the PexsC and PpcrG promoter regions. PexsC (A) or PpcrG (B) promoter probes labeled with 32P on the forward or reverse strand were treated with hydroxyl radicals, incubated with ExsAHis, and subjected to nondenaturing electrophoresis in a typical EMSA reaction. The unbound band and the ExsA-bound band corresponding to shift product 2 (representing ExsA bound to both sites 1 and 2) were eluted from the gel, separated by denaturing electrophoresis, and visualized by phosphorimaging. Maxam-Gilbert sequencing ladders (A+G) were included for orientation. The locations of the GnC and TGnnA sequences in binding site 1 and binding site 2 are indicated. (C) Summary of the footprinting data for the forward and reverse strands. The conserved GnC and TGnnA sequences (bold with larger typeface), adenine-rich region (gray box), and positions of ExsA binding sites 1 and 2 are indicated. Nucleotides indicated with arrows are those present in the unbound samples and required for maximal ExsA binding. (D) Proposed model for the binding of ExsA to site 1 in the PexoT promoter region. The amino acid residues that are proposed to contribute to base-specific recognition of the coding or noncoding strand of the PexoT promoter region are indicated.