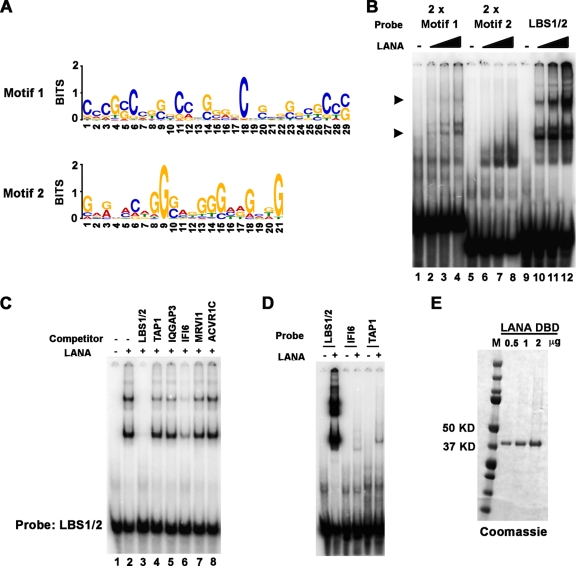
Fig 4.
Consensus binding site of LANA. (A) LANA consensus recognition sequences identified by both ChIP-Seq replicates were identified by MEME analysis and represented in Web Logo format. (B) EMSA analysis of 32P-labeled probes containing 2× motif 1 (lanes 1 to 4), 2× motif 2 (lanes 5 to 8), or LBS1/2 (lanes 9 to 12). Various amounts of LANA DBD protein was used in binding reaction (0, 0.2, 0.6, and 1.8 μg). Arrowheads indicate LANA-specific bound complexes. (C) EMSA with LANA bound to 32P-labeled LBS1/2 and challenged with a 1,000-fold excess of cold competitor oligonucleotides that included LBS1/2 (lane 3), the LANA peak sites in TAP1 (lane 4), IQGAP3 (lane 5), IFI6 (lane 6), MRVI1 (lane 7), and ACVR1C (lane 8). Lane 1 is probe only, and lane 2 is no competitor control. (D) EMSA analysis of 32P-labeled probes containing LBS1/2 or the LANA peak sites in IFI6 or TAP1. (E) Purified LANA-DBD shown on a Coomassie blue-stained gel.