Fig 4.
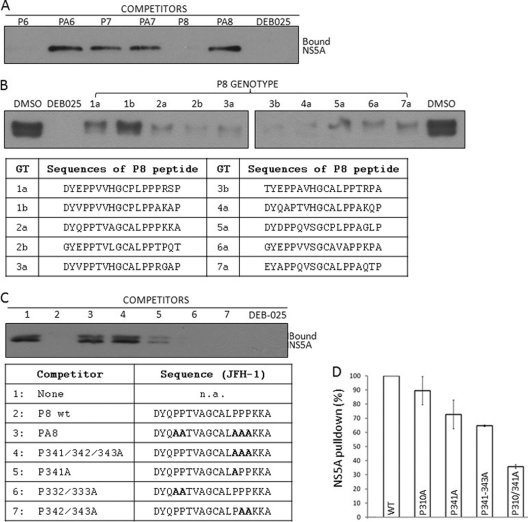
P6 and P8 represent a tandem CyPA-binding site of HCV NS5A. (A) Soluble P6, P7, and P8 peptides were evaluated for their ability to compete with NS5A from JFH-1-infected cell lysates for CyPA binding. The indicated peptides and DEB-025 were added to His-CyPA and incubated before NS5A-containing lysate was added to the pulldown assay. (B) Soluble P8 peptides from all seven genotypes were capable of binding to CyPA in the competition assay. Peptides were used at a 3.3-fold molar excess of the His-CyPA. (C) Proline at position 341 is the major contributor to CyPA binding by the JFH-1 P8 peptide. (D) P310 and P341 are the principal CyPA-binding sites on NS5A. HCV RNA containing NS5A of wt, P310A, P341A, P341-343A, or P310A P341A were expressed in Huh-7.5 cells using a recombinant vaccinia virus system and tested in the His-CyPA pulldown assay. The image shown is representative of two independent experiments. Note that in experiments where the two forms of phospho-NS5A (p56 and p58) were resolved by SDS-PAGE, CyPA binding was not biased toward either form, in agreement with previous results (63).