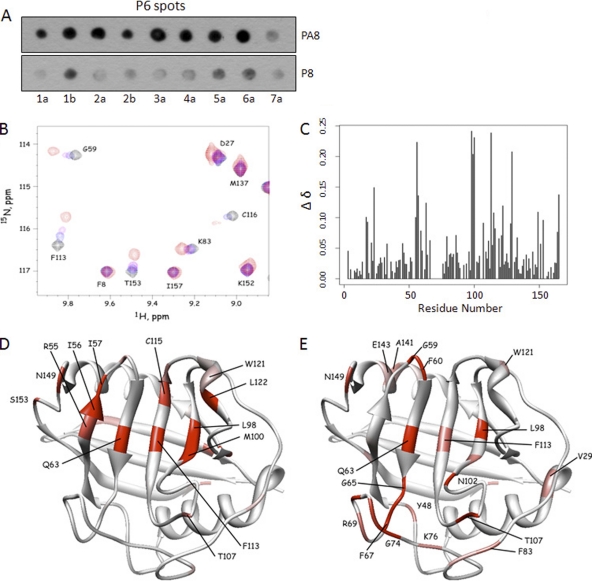
Fig 5.
P6 and P8 bind to the ligand-binding pocket of CyPA with similar binding strengths. (A) Soluble P8 peptide competed with P6 for CyPA binding in a proline-dependent manner. Soluble P8/PA8 peptides were included in approximately equal amounts as the total P6 peptides on the membrane (∼5 nmol per spot) in the spot-binding assay that measured P6-CyPA interaction. CyPA binding to P6 spots was reduced by the inclusion of the P8 but not the PA8 peptide. (B) NMR spectra showing the effect of P8 titration on the chemical shifts. Some resonances are insensitive to P8, while others show high sensitivity in both the 1H and 15N chemical shifts. (C) Chemical shift changes (Δδ) were quantified and plotted for each residue of CyPA. (D) Residues that exhibited chemical shift perturbations were highlighted in a structure of CyPA complexed with a fragment of the HIV-1 Gag protein (PDB access code 1FGL). Normalized Δδ values for P8 titration (65) were used to generate the color scheme: red, 0.75 < Δδ < 1; pink, 0.5 < Δδ < 0.74; white, Δδ < 0.50. (E) Residues with normalized Δδ values for P6 titration are shown in the same structure of CyPA. The color scheme is the same as that described for panel C.