Fig 2.
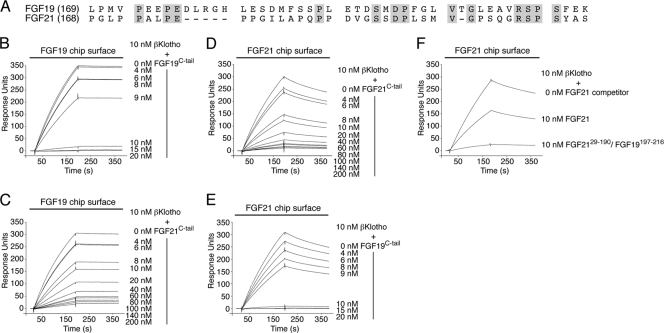
The C-terminal tail peptides of FGF19 and FGF21 bind to a common site on βKlotho, albeit with different affinities. (A) Alignment of the C-terminal tail sequences of human FGF19 and FGF21. Residue numbers are in parentheses to the left of the alignment. Gaps (dashes) were introduced to optimize the sequence alignment. Residues that are identical between human FGF19 and FGF21 are shaded gray. Note that the greatest degree of sequence identity (40%) is confined to the 20 most C-terminal residues. (B and C) Overlays of SPR sensorgrams illustrating inhibition by FGF19C-tail (B) or FGF21C-tail (C) of βKlotho binding to FGF19. FGF19 was immobilized on a biosensor chip, and mixtures of a fixed concentration of βKlotho ectodomain with increasing concentrations of either FGF19C-tail or FGF21C-tail were passed over the chip. (D and E) Overlays of SPR sensorgrams illustrating inhibition by FGF21C-tail (D) or FGF19C-tail (E) of βKlotho binding to FGF21. FGF21 was immobilized on a biosensor chip, and mixtures of a fixed concentration of βKlotho ectodomain with increasing concentrations of either FGF19C-tail or FGF21C-tail were passed over the chip. Note that FGF19C-tail is more potent than FGF21C-tail at inhibiting βKlotho binding to FGF19 or FGF21 (compare panels B and E with panels C and D). (F) Overlay of SPR sensorgrams illustrating inhibition by FGF2129–190/FGF19197–216 or FGF21 of βKlotho binding to FGF21 immobilized on a biosensor chip. βKlotho ectodomain alone and 1:1 mixtures of βKlotho ectodomain with either FGF2129–190/FGF19197–216 or FGF21 were passed over a FGF21 chip. Note that the FGF2129–190/FGF19197–216 chimera is a more potent competitor for βKlotho binding than is native FGF21. The data shown in figure panels B to F are representative of two to three independent experiments.