Fig 7.
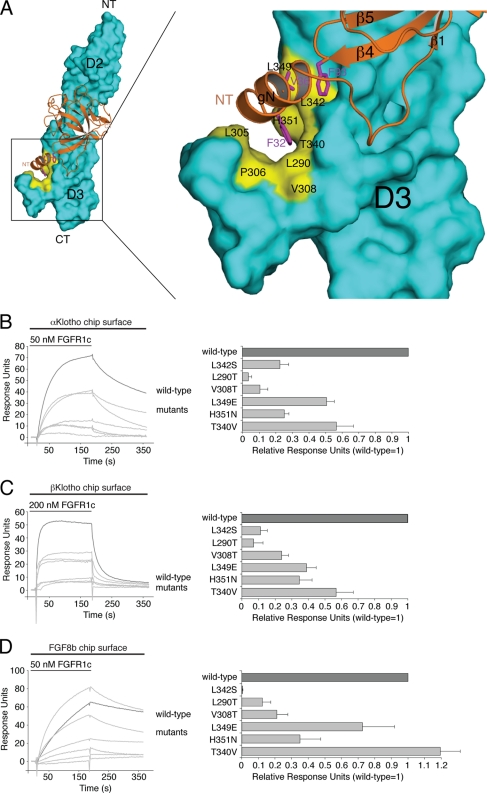
Klotho proteins and FGF8b engage the same hydrophobic groove in D3 of FGFR1c. (A) Structural model of the FGF8b-FGFR1c complex. The model was created by superimposing the ligand-binding domain of FGFR1c from the FGF2-FGFR1c crystal structure (PDB ID, 1CVS [55]) onto the ligand-binding domain of FGFR2c in the FGF8b-FGFR2c crystal structure (PDB ID, 2FDB [50]). On the left is a view of the whole model, and on the right is a close-up view of the ligand-receptor D3 interface. FGF8b is shown as a ribbon diagram, and FGFR1c is shown as a space-filling molecular surface. Note that F32 and V36 of the N-terminal g helix and F93 of the β4-β5 loop of FGF8b bind to a hydrophobic groove in the D3 domain of FGFR1c formed by L290, L305, P306, V308, T340, L342, L349, and H351. NT and CT denote N and C termini of FGF8b and FGFR1c, respectively. (B, C, and D) Overlays of SPR sensorgrams illustrating binding of wild-type and mutant FGFR1c proteins to αKlotho (B), βKlotho (C), and FGF8b (D). αKlotho ectodomain, βKlotho ectodomain, or FGF8b was immobilized on biosensor chips, and increasing concentrations of either wild-type or mutant FGFR1c ligand-binding domain were passed over the chips. Maximal binding responses of FGFR1c mutants relative to wild-type protein were plotted (25). As we have previously reported (53), the L342S mutation causes a major loss in binding affinity of FGFR1c for FGF8b.