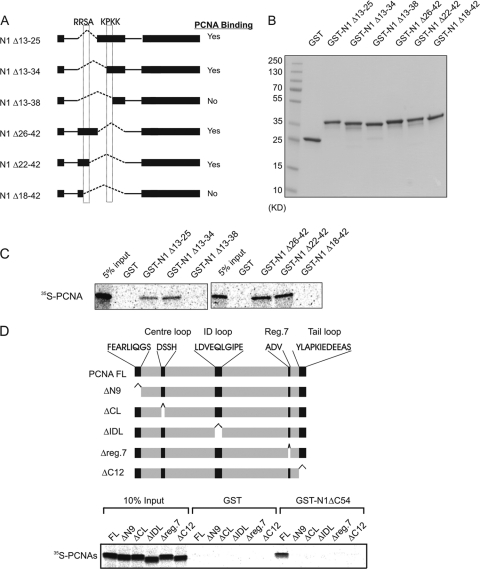
Fig 2.
Identification of the PCNA-binding regions in HMGN1. (A) Schematic representation of HMGN1 deletion mutants for fine mapping of the region responsible for interaction with PCNA. The dotted boxes depict the locations of the critical HMGN1 tetrapeptides necessary for PCNA binding. (B) SDS-PAGE analysis of purified deletion mutants shown in panel A. (C) Autoradioagraphic identification of HMGN1 regions that interact with PCNA using cell-free 35S labeling and a GST pulldown assay. (D) Identification of PCNA deletions which abrogate the interaction between PCNA and HMGN1. 35S-labeled PCNA or PCNA deletion mutants were incubated with either purified ΔC54 HMGN1-GST or only GST proteins, and the complex was purified on glutathione-Sepharose, fractionated on an SDS-PAGE gel, and visualized by autoradiography.