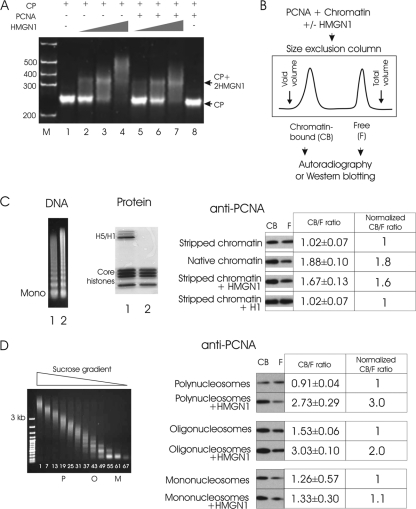
Fig 3.
HMGN1 facilitates PCNA binding to chromatin in vitro. (A) HMGN1 does not facilitate the binding of PCNA to the 147-bp CP. A mobility shift assay demonstrates that HMGN1 (lanes 2 to 4) but not PCNA (lane 8) binds to CPs. The lack of supershifts in lanes 5 to 7 indicates that HMGN1 does not promote the binding of PCNA to CPs. (B) Size exclusion chromatography assay for analysis of the effect of HMGN1 on the binding of PCNA to chromatin. PCNA is added to chromatin in either the presence or absence of HMGN1. Chromatin-bound PCNA elutes in the void volume, while free PCNA elutes in the total volume. (C) HMGN1 enhances the binding of PCNA to chromatin. Left side, DNA and protein gels of micrococcal nuclease-digested chromatin. Lane 1, not salt stripped; lane 2, stripped with 0.5 M NaCl. Right side, Western analysis of free (F) and chromatin-bound (CB) PCNA present in various chromatin preparations. The ratio of CB to F PCNA, determined by scanning the film, is indicated. An average of three replicates is presented, with standard deviations. Note that addition of HMGN1, but not of H1, to chromatin increased the amount of chromatin-bound PCNA. (D) HMGN1 preferentially enhances PCNA binding to longer chromatin fragments. Left, sucrose gradient fractionation of salt-stripped micrococcal nuclease-digested chromatin; right, Western analysis of the relative amounts of chromatin-bound and free PCNA. PCNA was added to either polynucleosomes (P, fragments ranging from 9 to 15 nucleosomes), oligonucleosomes (O, 2 to 6 nucleosomes), or mononucleosomes (M) in either the presence or absence of HMGN1 and fractionated on size exclusion columns (see panel B).