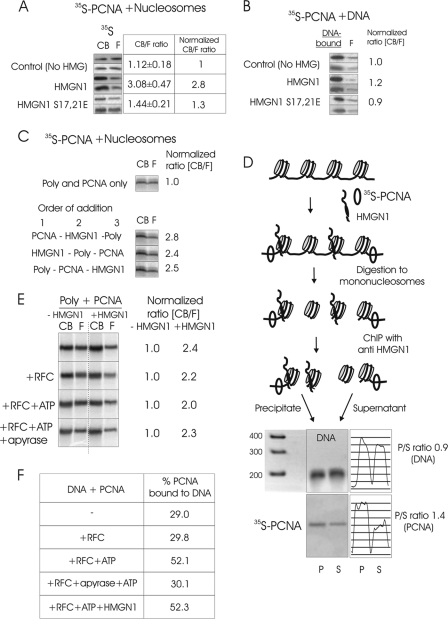
Fig 4.
(A) The interaction of HMGN1 with nucleosomes enhances the binding of PCNA to chromatin. 35S-PCNA was added to salt-stripped polynucleosomes in the presence of either native HMGN1 or mutant HMGN1 which does not bind to chromatin, and the mixture was fractionated on size exclusion columns. The relative amounts of chromatin-bound (CB) and free (F) PCNA were determined by ImageQuant analyses of autoradioagraphic images. Shown are images from 2 separate experiments. The CB/F values were obtained from triplicate experiments. (B) HMGN1 does not affect the binding of PCNA to purified DNA. (C) HMGN1 improves the binding of PCNA to polynucleosome (poly) independently of order of addition. (D) PCNA binds preferentially adjacent to HMGN-containing nucleosomes. Mixtures of polynucleosomes, HMGN1, and 35S-PCNA were formaldehyde cross-linked, digested to mononucleosomes, and immunoprecipitated with anti-HMGN1. The amount of 35S-PCNA in the precipitate (HMGN-containing nucleosomes) and supernatant was analyzed by polyacrylamide gels followed by autoradiography. Scans of the DNA gel and of the autoradiograph, and the ratio of DNA and PCNA in the precipitate (P) to that in the supernatant (S), are on the right. (E) HMGN1-mediated enhancement of PCNA to polynucleosomes is not dependent on RFC. Shown are autoradioagraphs of 35S-PCNA in the CB and F fractions in the presence and absence of HMGN1. The RFC, ATP, and apyrase present in the various experiments are indicated on the left. The ratio of CB/F for 35S-PCNA in the absence of HMGN1 was normalized to 1.0. (F) HMGN1 does not affect the RFC-mediated loading of PCNA onto purified DNA.