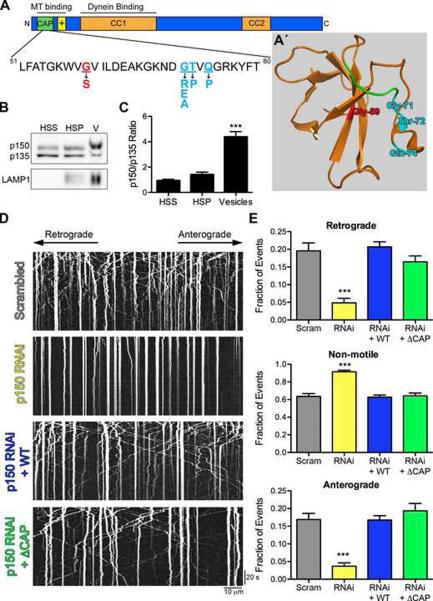
Figure 1. The highly conserved CAP-Gly domain of p150Glued is not required for axonal transport of lysosomes.
(A) Schematic of the p150Glued subunit of dynactin. The CAP-Gly domain (green) binds both MTs and MT plus-end binding proteins. The basic domain (yellow) also independently binds MTs. The HMN7B (red) and the Perry syndrome (cyan) point mutations are shown below. (A′) Crystal structure of the CAP-Gly domain of p150Glued modified from (Honnappa et al., 2006) (PDB ID 2HKQ). The GKNDG motif is in green and the disease-associated mutations are colored as in (A). The HMN7B mutation is buried at the core of the CAP-Gly domain and may disrupt protein folding, while the Perry syndrome mutations are surface-exposed and may predominantly interfere with protein-protein interactions. Also see Movie S1. (B) Purified vesicles (V) from mouse brain enriched for LAMP1 were probed for p150Glued using a polyclonal antibody that recognizes both the full-length and p135 isoforms. (C) Purified vesicles had an increased ratio of full-length p150Glued to p135 compared to the high speed supernatant (HSS) and high-speed pellet (HSP) fractions. Mean ± SEM, n=3 vesicle purifications, ***P<0.001 compared to HSS, one-way ANOVA Bonferroni post test. (D) Kymographs of LAMP1-RFP motility in primary dorsal root ganglion (DRG) neurons imaged at 4 days in vitro (DIV) after transfection with two scrambled siRNAs or two siRNAs to p150Glued. siRNAs were also transfected with wild-type full-length p150Glued (WT) or ΔCAP-Gly p150Glued (ΔCAP) resistant to the siRNA. Kymographs drawn along neurite processes represent movement over time so motile organelles appear as diagonal lines while paused organelles appear as vertical lines. Images were acquired at 366 ms per frame for 2.2 minutes; scale bars for the x and y-axes represent 10 μm and 20 seconds, respectively. Kymographs show the first 300 frames, full movies shown in Movie S2. (E) Lysosomal motility was quantified from the kymographs. Depletion of p150Glued significantly disrupted anterograde and retrograde motility and caused a corresponding increase in non-motile events. Expression of either wild-type or ⊗CAP-Gly p150Glued rescued this disruption. >800 vesicles were counted per condition. Mean ± SEM, n=12–15 neurons per condition, ***P<0.001 compared to scrambled siRNAs, one-way ANOVA Bonferroni post test. Also see Figure S1.