Abstract
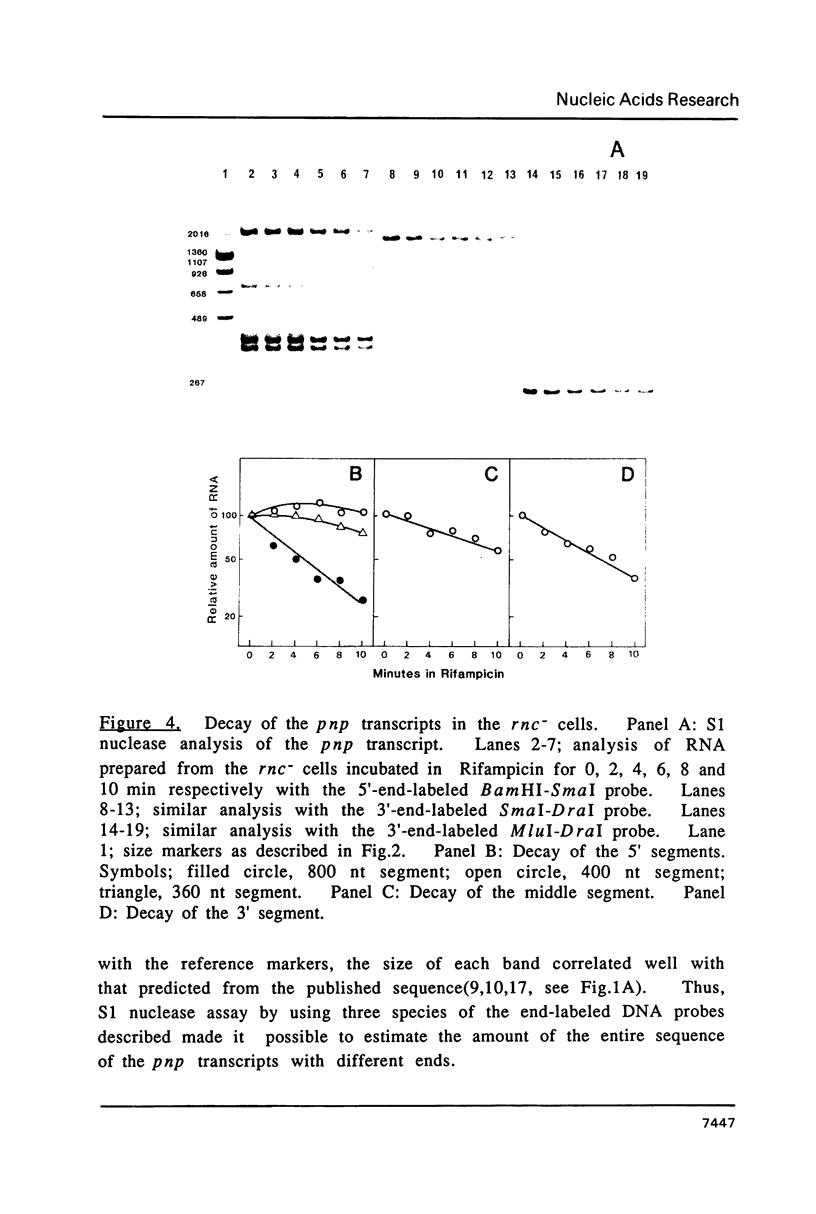
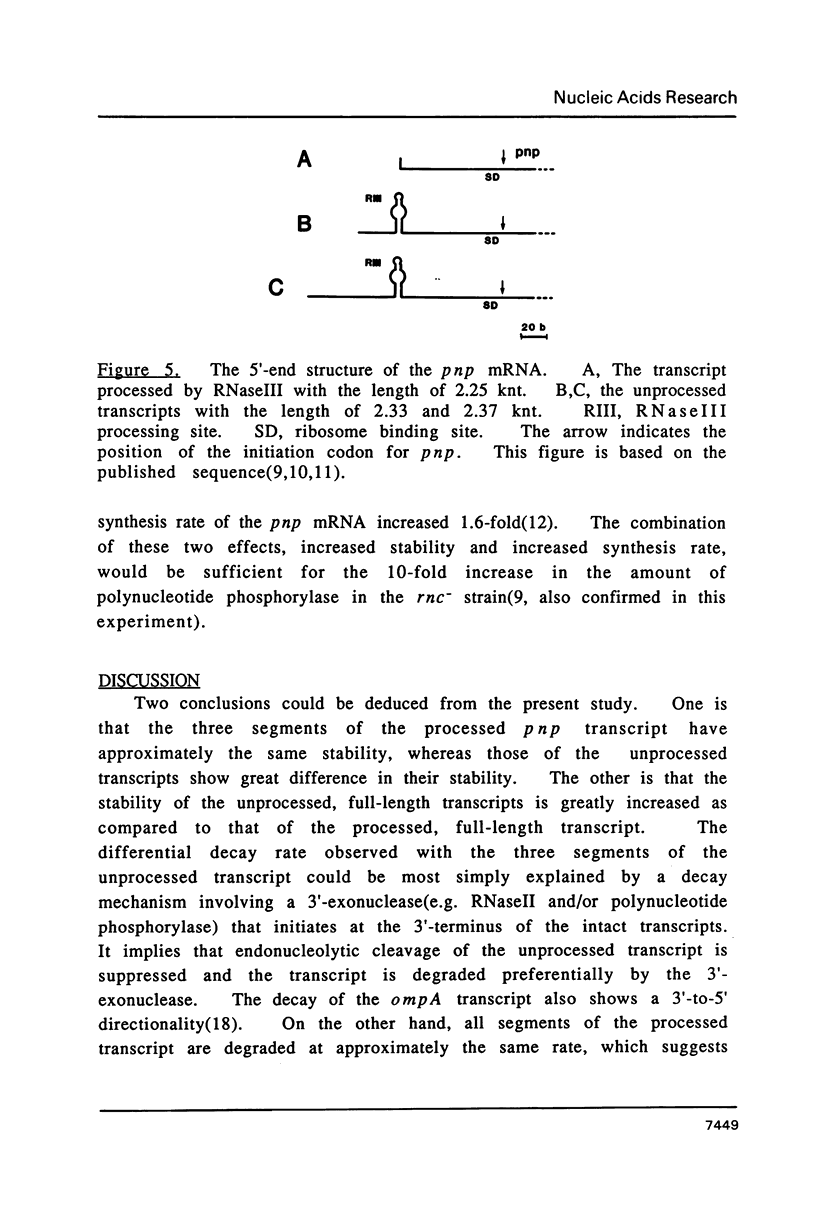
The transcript covering pnp, the gene encoding polynucleotide phosphorylase, is processed by RNaseIII at the 5'-upstream site of the pnp gene. In the RNaseIII-deficient strain, three species of the unprocessed transcript with different lengths could be detected. In this study, the stability of each transcript was analyzed by SI nuclease protection assay. The results show that the half-lives of the unprocessed transcripts are 8 min, whereas the half-life of the processed transcript is 1.5 min. It is also shown that the 5' segment of the unprocessed transcripts is more stable than the middle or the 3' segment.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Apirion D., Watson N. Mapping and characterization of a mutation in Escherichia coli that reduces the level of ribonuclease III specific for double-stranded ribonucleic acid. J Bacteriol. 1975 Oct;124(1):317–324. doi: 10.1128/jb.124.1.317-324.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. G., Higgins C. F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988 Dec 10;72(1-2):15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Blumer K. J., Steege D. A. mRNA processing in Escherichia coli: an activity encoded by the host processes bacteriophage f1 mRNAs. Nucleic Acids Res. 1984 Feb 24;12(4):1847–1861. doi: 10.1093/nar/12.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton Z. F., Gross C. A., Watanabe K. K., Burgess R. R. The operon that encodes the sigma subunit of RNA polymerase also encodes ribosomal protein S21 and DNA primase in E. coli K12. Cell. 1983 Feb;32(2):335–349. doi: 10.1016/0092-8674(83)90453-1. [DOI] [PubMed] [Google Scholar]
- Båga M., Göransson M., Normark S., Uhlin B. E. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell. 1988 Jan 29;52(2):197–206. doi: 10.1016/0092-8674(88)90508-9. [DOI] [PubMed] [Google Scholar]
- Cole J. R., Nomura M. Changes in the half-life of ribosomal protein messenger RNA caused by translational repression. J Mol Biol. 1986 Apr 5;188(3):383–392. doi: 10.1016/0022-2836(86)90162-2. [DOI] [PubMed] [Google Scholar]
- Kurihara T., Nakamura Y. Cloning of the nusA gene of Escherichia coli. Mol Gen Genet. 1983;190(2):189–195. doi: 10.1007/BF00330639. [DOI] [PubMed] [Google Scholar]
- Melefors O., von Gabain A. Site-specific endonucleolytic cleavages and the regulation of stability of E. coli ompA mRNA. Cell. 1988 Mar 25;52(6):893–901. doi: 10.1016/0092-8674(88)90431-x. [DOI] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Higgins C. F. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell. 1987 Dec 24;51(6):1131–1143. doi: 10.1016/0092-8674(87)90599-x. [DOI] [PubMed] [Google Scholar]
- Nilsson G., Belasco J. G., Cohen S. N., von Gabain A. Effect of premature termination of translation on mRNA stability depends on the site of ribosome release. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4890–4894. doi: 10.1073/pnas.84.14.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson G., Belasco J. G., Cohen S. N., von Gabain A. Growth-rate dependent regulation of mRNA stability in Escherichia coli. Nature. 1984 Nov 1;312(5989):75–77. doi: 10.1038/312075a0. [DOI] [PubMed] [Google Scholar]
- Portier C., Dondon L., Grunberg-Manago M., Régnier P. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5' end. EMBO J. 1987 Jul;6(7):2165–2170. doi: 10.1002/j.1460-2075.1987.tb02484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Régnier P., Grunberg-Manago M., Portier C. Nucleotide sequence of the pnp gene of Escherichia coli encoding polynucleotide phosphorylase. Homology of the primary structure of the protein with the RNA-binding domain of ribosomal protein S1. J Biol Chem. 1987 Jan 5;262(1):63–68. [PubMed] [Google Scholar]
- Régnier P., Portier C. Initiation, attenuation and RNase III processing of transcripts from the Escherichia coli operon encoding ribosomal protein S15 and polynucleotide phosphorylase. J Mol Biol. 1986 Jan 5;187(1):23–32. doi: 10.1016/0022-2836(86)90403-1. [DOI] [PubMed] [Google Scholar]
- Salser W., Gesteland R. F., Bolle A. In vitro synthesis of bacteriophage lysozyme. Nature. 1967 Aug 5;215(5101):588–591. doi: 10.1038/215588a0. [DOI] [PubMed] [Google Scholar]
- Takata R., Mukai T., Aoyagi M., Hori K. Nucleotide sequence of the gene for Escherichia coli ribosomal protein S15 (rpsO). Mol Gen Genet. 1984;197(2):225–229. doi: 10.1007/BF00330967. [DOI] [PubMed] [Google Scholar]
- Takata R., Mukai T., Hori K. Attenuation and processing of RNA from the rpsO-pnp transcription unit of Escherichia coli. Nucleic Acids Res. 1985 Oct 25;13(20):7289–7297. doi: 10.1093/nar/13.20.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata R., Mukai T., Hori K. RNA processing by RNase III is involved in the synthesis of Escherichia coli polynucleotide phosphorylase. Mol Gen Genet. 1987 Aug;209(1):28–32. doi: 10.1007/BF00329832. [DOI] [PubMed] [Google Scholar]
- von Gabain A., Belasco J. G., Schottel J. L., Chang A. C., Cohen S. N. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci U S A. 1983 Feb;80(3):653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]





