Fig. 1.
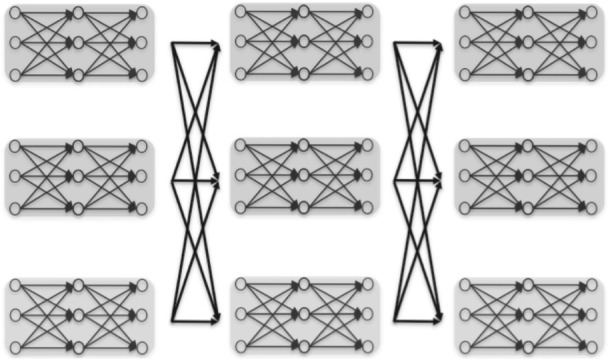
Schematic structure of our model (haploid version is displayed for simplicity) over nine SNPs with three windows each of length three SNPs. In each window, the haplotypes of each ancestral population are modeled using distinct HMMs (denoted in different colors). Transitions that change the ancestral population are allowed only at the boundary of consecutive windows. This framework is generic in that any model [e.g. Li and Stephens (Li and Stephens, 2003), fastPHASE (Scheet and Stephens, 2006)] can be used to account for the ancestral LD.
