Abstract
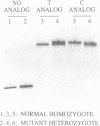
A simple primer extension assay has been developed to distinguish homologous DNA segments differing by as little as a single nucleotide. DNA strands are synthesized with one of the four natural nucleotides replaced with an analog that affects electrophoretic mobility. DNAs that are the same length but differ in the number of analog molecules per strand exhibit different mobilities on a sequencing gel. In combination with the polymerase chain reaction (PCR; 1, 2), this method has been used to distinguish mutant and normal alleles of the human insulin receptor gene that differ by a single-base substitution. The method appears to be generally applicable to the detection of any nucleotide polymorphism in any segment of DNA.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Botstein D., White R. L., Skolnick M., Davis R. W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980 May;32(3):314–331. [PMC free article] [PubMed] [Google Scholar]
- Caskey C. T. Disease diagnosis by recombinant DNA methods. Science. 1987 Jun 5;236(4806):1223–1229. doi: 10.1126/science.3296189. [DOI] [PubMed] [Google Scholar]
- Cotton R. G., Rodrigues N. R., Campbell R. D. Reactivity of cytosine and thymine in single-base-pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4397–4401. doi: 10.1073/pnas.85.12.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke D. R., Hoener P. A., Collins F. S. Direct sequencing of enzymatically amplified human genomic DNA. Proc Natl Acad Sci U S A. 1988 Jan;85(2):544–548. doi: 10.1073/pnas.85.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S. G., Lerman L. S. DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with melting theory. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1579–1583. doi: 10.1073/pnas.80.6.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T., Bevins C. L., Cama A., Ojamaa K., Marcus-Samuels B., Kadowaki H., Beitz L., McKeon C., Taylor S. I. Two mutant alleles of the insulin receptor gene in a patient with extreme insulin resistance. Science. 1988 May 6;240(4853):787–790. doi: 10.1126/science.2834824. [DOI] [PubMed] [Google Scholar]
- Landegren U., Kaiser R., Caskey C. T., Hood L. DNA diagnostics--molecular techniques and automation. Science. 1988 Oct 14;242(4876):229–237. doi: 10.1126/science.3051381. [DOI] [PubMed] [Google Scholar]
- Landegren U., Kaiser R., Sanders J., Hood L. A ligase-mediated gene detection technique. Science. 1988 Aug 26;241(4869):1077–1080. doi: 10.1126/science.3413476. [DOI] [PubMed] [Google Scholar]
- Langer P. R., Waldrop A. A., Ward D. C. Enzymatic synthesis of biotin-labeled polynucleotides: novel nucleic acid affinity probes. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6633–6637. doi: 10.1073/pnas.78.11.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullis K. B., Faloona F. A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Larin Z., Maniatis T. Detection of single base substitutions by ribonuclease cleavage at mismatches in RNA:DNA duplexes. Science. 1985 Dec 13;230(4731):1242–1246. doi: 10.1126/science.4071043. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Lumelsky N., Lerman L. S., Maniatis T. Detection of single base substitutions in total genomic DNA. Nature. 1985 Feb 7;313(6002):495–498. doi: 10.1038/313495a0. [DOI] [PubMed] [Google Scholar]
- Novack D. F., Casna N. J., Fischer S. G., Ford J. P. Detection of single base-pair mismatches in DNA by chemical modification followed by electrophoresis in 15% polyacrylamide gel. Proc Natl Acad Sci U S A. 1986 Feb;83(3):586–590. doi: 10.1073/pnas.83.3.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prober J. M., Trainor G. L., Dam R. J., Hobbs F. W., Robertson C. W., Zagursky R. J., Cocuzza A. J., Jensen M. A., Baumeister K. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science. 1987 Oct 16;238(4825):336–341. doi: 10.1126/science.2443975. [DOI] [PubMed] [Google Scholar]
- Päbo S., Wilson A. C. Polymerase chain reaction reveals cloning artefacts. Nature. 1988 Aug 4;334(6181):387–388. doi: 10.1038/334387b0. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sheffield V. C., Cox D. R., Lerman L. S., Myers R. M. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci U S A. 1989 Jan;86(1):232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T. E., Rhodes C., Rigby P. W., Berg P. Biochemical method for mapping mutational alterations in DNA with S1 nuclease: the location of deletions and temperature-sensitive mutations in simian virus 40. Proc Natl Acad Sci U S A. 1975 Mar;72(3):989–993. doi: 10.1073/pnas.72.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich A., Bell J. R., Chen E. Y., Herrera R., Petruzzelli L. M., Dull T. J., Gray A., Coussens L., Liao Y. C., Tsubokawa M. Human insulin receptor and its relationship to the tyrosine kinase family of oncogenes. 1985 Feb 28-Mar 6Nature. 313(6005):756–761. doi: 10.1038/313756a0. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Johnson M. J., Hirose T., Miyake T., Kawashima E. H., Itakura K. The use of synthetic oligonucleotides as hybridization probes. II. Hybridization of oligonucleotides of mixed sequence to rabbit beta-globin DNA. Nucleic Acids Res. 1981 Feb 25;9(4):879–894. doi: 10.1093/nar/9.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]