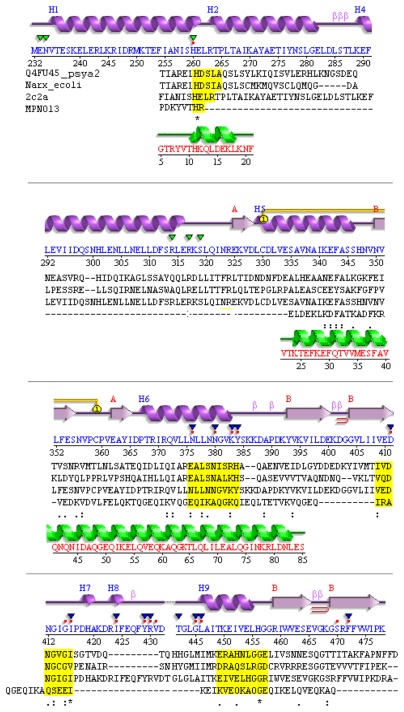
Figure 1.
Divergent TCS sensor in M. pneumoniae.
Notes: Compared are the structure template (T. maritima), structure of NarX from E. coli, P. arcticus, and MPN013 (M. pneumoniae). Aligned are the secondary structure from PDB template 2c2a_A (top, magenta; HK853 from T. maritima) and its sequence (blue), valid (sequences aligned) for NarX from P. arcticus and the sequence of MPN013. Conserved residues are highlighted by yellow boxes. Below the secondary structure triangles indicate binding sites annotated in PDBSum (green: ADP binding site, blue SO4 binding site, red dots ligand binding site). Conserved residues for TCS (see above) are highlighted in yellow boxes. Structure: Calculated secondary structure (green) according to the SWISS-MODEL template for MPN013 (PDB entry 2ba2_A for MPN010).