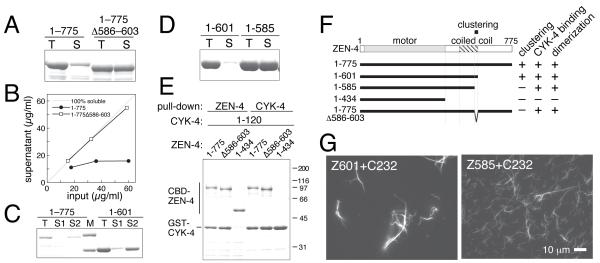
Figure 2.
In vitro characterization of the clustering of C. elegans centralspindlin. (A-D) Bacterially expressed ZEN-4 proteins were purified in the presence of 250 mM salt and diluted to (A, B) 150 mM or (C, D) 83 mM. Total input (T) and supernatant (S) after centrifugation were analysed by SDS-PAGE. For C, after the first supernatant (S1) was removed, the precipitate (not shown) was resuspended into a high salt buffer and centrifuged to obtain the second supernatant (S2). (E) CYK-4 binding was assayed by reciprocal pull-down from E. coli lysates expressing both ZEN-4 fragments tagged with chitin-binding domain (CBD) and CYK-4 1-120 tagged with glutathione S-transferase (GST). (F) Schematic model of ZEN-4 and deletion constructs used in this study with a summary of their in vitro properties. ZEN-4 is comprised of a motor domain, a neck region containing several helix-breaking proline residues, a 100 amino acid region that forms a parallel coiled coil [20] and a tail domain. A small region (‘clustering element’) is required for clustering. (G) Microtubule-bundling assays with ZEN-4 constructs competent (Z601) or incompetent (Z585) for clustering complexed with CYK-4 1-232 (C232).