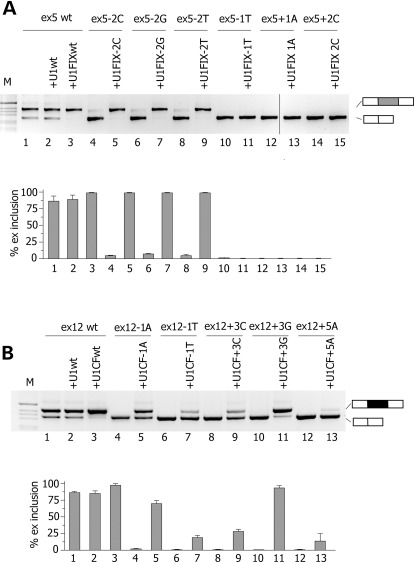
Figure 2.
Effect of increased complementarity of U1 snRNAs to normal and mutated donor splice site on pre-mRNA splicing. (A) Effect of complementary U1 snRNA to the F9 exon 5 5′ss. The upper panel shows the analysis of spliced transcripts. Minigenes were transfected in HeLa cells with the empty vector (lanes 1, 4, 6, 8, 10, 12, 14) or with modified U1s (lanes 3, 5, 7, 9, 11, 13, 15) and splicing pattern evaluated by RT–PCR. Amplified products were resolved on a 2% agarose gel. The identity of the U1 snRNAs is shown in Supplementary Material, Figure S2. Lower panel is the quantification of exon 5 splicing pattern after co-transfection with modified U1 snRNAs. Data are expressed as means ± SD of three independent experiments done in duplicate. (B) Effect of complementary U1 snRNAs to the CFTR exon 12 5′ss. Minigenes were transfected in HeLa cells with the empty vector (lanes 4, 6, 8) or with modified U1s (lanes 3, 5, 7, 9) and the splicing pattern evaluated by RT–PCR. Amplified products were resolved on a 2% agarose gel. The identity of the U1 snRNAs is shown in Supplementary Material, Figure S2. Lower panel is the quantification of exon 12 splicing pattern. Data are expressed as means ± SD of three independent experiments done in duplicate.