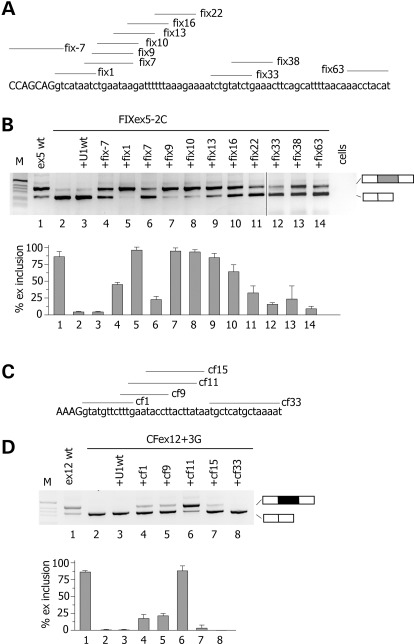
Figure 3.
Identification of ExSpeU1s in F9 exon 5 and CFTR exon 12. (A and C) Schematic representation of binding sites of the ExSpeU1s in F9 exon 5 and CFTR exon 12, respectively. Donor site and downstream intronic region are shown and exonic and intronic sequences are in upper and lower case, respectively. Lines correspond to the target-binding sequences of ExSpeU1s U1 snRNAs on the nascent pre-mRNA. (B) Analysis of F9 exon 5-spliced transcripts. The FIXex5-2C mutant minigene was transfected in HeLa cells alone (lane 1), with the empty vector (lane 2), or with plasmids encoding for the different ExSpeU1s. The pattern of splicing was evaluated by RT–PCR and amplified products were resolved on a 2% agarose gel. Lower panel is the quantification of exon 5 splicing pattern. (D) Analysis of CFTR exon 12-spliced transcripts. The mutant ex12 + 3G minigene was transfected in HeLa cells alone (lane 1), with the empty vector (lane 2), or with plasmids encoding for the ExSpeU1s. The pattern of splicing was evaluated by RT–PCR and amplified products were resolved on a 2% agarose gel. Lower panel is the quantification of exon 5 splicing pattern.