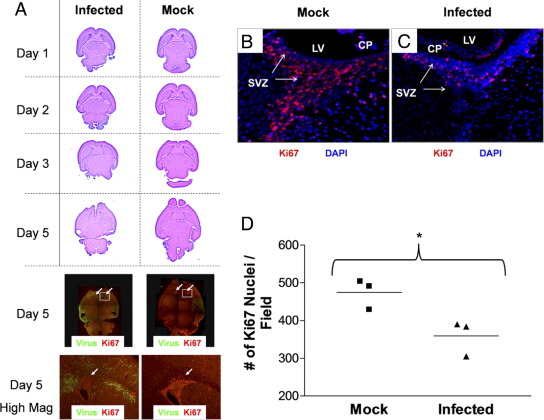
Figure 1.
Lethal inoculum of eGFP-CVB3 during the neonatal period hinders CNS development. One-day-old pups were infected with eGFP-CVB3 (2 × 106 pfu i.c.) or were mock infected. Brains were harvested and fixed in 10% neutral buffered formalin at the indicated time points PI. Paraffin sections were stained by H&E or immunostained for the cellular proliferation antigen (Ki-67, red) and viral protein expression (GFP, green) and observed by fluorescence microscopy. A: Two color channels were merged into a single image (original magnification, ×5) and composites of overlapping fields were assembled to illustrate a complete transverse section of the neonatal CNS for two representative mice. Normal CNS development was observed in mock-infected pups. As would be expected for regions of neurogenesis in the CNS, a large number of Ki-67+ cells were observed in the SVZ near the lateral ventricles and in the cerebellum. In pups infected with eGFP-CVB3 and harvested 5 days later, the overall size of the brain was significantly smaller and the ventricles were severely distended. Furthermore, CNS lesions were seen in the hippocampus and cortex regions, in parallel with high levels of viral protein expression. The cerebellum (only partly visible) had little to no detectable viral protein levels and displayed similar numbers of proliferating (Ki-67+) cells compared with mock-infected control mice. Higher magnification for day 5 fluorescent images revealed that the number of proliferating (Ki-67+) cells was substantially reduced in the SVZ (white arrow) near the lateral ventricles, as compared with mock-infected control mice. B: High levels of Ki-67+ signal were observed in the SVZ of mock-infected mice. C: In contrast, the level of Ki-67+ signal was sharply reduced within infected mice. D: Quantification of Ki-67+ signal in the SVZ showed substantially reduced levels of staining in the SVZ of infected mice (*P = 0.0327). Quantification of Ki-67+ cells was done in triplicate (three mice per group and average of three fields per mouse) using ImageJ software as described previously.10