Abstract
Cocaine esterase (CocE) is the most efficient cocaine-metabolizing enzyme tested in vivo to date, displaying a rapid clearance of cocaine and a robust protection against cocaine’s toxicity. Two potential obstacles to the clinical application of CocE, however, lie in its proteolytic degradation and induced immune response. To minimize these potential obstacles, we attempted non-disruptive cell encapsulation by creating a cell permeable form of CocE, which was achieved by covalently linking a thermally stable CocE mutant (dmCocE) with cell penetrating peptides (CPPs). Two types of CPPs, Tat and the low molecular weight protamine (LMWP), were used in this study. Two types of disulfide-bridged chemical conjugates, Tat-S-S-dmCocE and LMWP-S-S-dmCocE, were synthesized and then purified by heparin affinity chromatography. In addition, four recombinant CPP-dmCocE fusion proteins, Tat-N-dmCocE, LMWP-N-dmCocE, dmCocE-C-Tat, and dmCocE-C-LMWP, were constructed, expressed in E. coli and purified as soluble proteins. Among these six CPP-dmCocE variants, LMWP-S-S-dmCocE showed the highest cocaine-hydrolyzing activity, and dmCocE-C-Tat had the highest production yield. To evaluate their cellular uptake behavior, a covalently-linked fluorophore (FITC) was utilized to visualize the cellular uptake of all six CPP-dmCocE variants in living HeLa cells. All the six variants exhibited cellular uptake, but their intracellular distribution phenotypes differed. While the chemical conjugates showed primarily cytoplasmic distribution, which was likely due to the reduction of the disulfide linkage between CPP and dmCocE, all the other four recombinant fusion proteins displayed both nuclear and cytoplasmic localization, with dmCocE-C-CPP exhibiting higher cytoplasmic distribution during cellular uptake. Based on a balanced consideration of essentials for clinical application, including parameters such as high cocaine-hydrolyzing efficiency, large production yield, major cytoplasmic distribution, etc., the dmCocE-C-Tat fusion protein seems to be the best candidate from this investigation. Further in vivo studies of the cell-encapsulated dmCocE-C-Tat in hydrolyzing cocaine and alleviating immunogenicity and proteolytic degradation in established, clinically relevant mouse models are currently underway in our laboratories. Findings from this research are not only useful for developing other new CPP-CocE constructs, but also valuable for establishing a non-disruptive cell-encapsulation technology for other protein therapeutics that are known to be immunogenic for direct clinical application.
Keywords: Cell penetrating peptides (CPP), cocaine esterase (CocE), Tat, low molecular weight protamine (LMWP)
Introduction
Cocaine is one of the most addictive and commonly abused illicit drugs1. In addition to its widespread prevalence and potential for addiction, cocaine is also a highly toxic substance that can be lethal and often requires immediate detoxification treatment2. Recently, bacterial cocaine esterase (CocE), a globular 574 amino-acid monomeric esterase isolated from Rhidococcus sp. MB1 in the rhizosphere soil of the cocaine-producing plant3, has been identified as the most efficient naturally-occurring enzyme for hydrolyzing cocaine to inactive metabolites4. Research on animal models has shown that CocE significantly reduced cocaine concentration in circulation and reversed cocaine toxicity5. In spite of encouraging results in animals, potential obstacles to the clinical application of CocE in humans lie in its thermal instability, proteolytic degradation, and induced immune response. Native CocE is rapidly deactivated at physiological temperature; its in vitro half-life at 37 °C is only ~13.7 min5. In addition, like most exogenous proteins, CocE is prone to be cleaved by serum proteases in circulation. Moreover, since CocE is a bacterial protein, it could trigger the production of anti-CocE antibodies that would not only deactivate CocE but be also potentially deleterious to the host. To improve its thermal stability, several mutants of CocE have been designed using a computational structural analysis, constructed and tested. These mutants successfully extended the in vitro half-life from minutes to several hours6, 7. However, the proteolytic degradation and induced immune response remain to be critical issues, which must be solved prior to any possible clinical application.
Aside from conventional strategies to reduce CocE’s proteolysis and immunogenicity by attaching polyethylene glycol (PEG) chains8 , another attractive method is encapsulating CocE into living cells. As long as CocE is confined inside the cells, it can be protected from proteolytic degradation and immune surveillance by the host. Indeed, cell encapsulation has been applied to a variety of therapeutic enzymes for the treatment of different diseases such as Gaucher’s disease, methanol intoxication, and acute lymphoblastic leukemia9–11. This strategy has successfully attenuated many inherent drawbacks in traditional enzyme replacement therapies, including short half-lives, immune and allergic reactions, and host toxicity12. Furthermore, owing to the fact that cocaine is a small lipophilic molecule, cocaine readily crosses cell membranes via simple diffusion and distributes to the cellular compartments. According to reports of intravenous cocaine injection in humans, cocaine rapidly accumulates in the brain as well as red blood cells (RBC) and reaches maximal concentration within a short period of <5 minutes13, 14. More importantly, the cocaine concentrations in brain and RBC have shown to remain approximately two-fold higher than the concentrations in plasma13, 14. This supports the concept of developing a cell-encapsulated form of CocE to avoid the proteolysis and immunogenicity issues, thereby achieving an effective yet non-toxic cocaine detoxification.
It is well-known that the cell membrane is only permeable to small (< 700 Da) lipophilic molecules but not to macromolecules including enzymes15. To solve this permeability issue for cell encapsulation, several approaches including hypo-osmosis and electroporation have been developed to load enzymes into living cells. Nevertheless, these approaches all require disruption of the cell membrane to enhance permeability. By generating large pores or perturbations on the cell membrane, these methods often lead to morphological alteration and physiological dysfunction of the cells12.
To achieve a non-disruptive cell encapsulation for CocE, we proposed an innovative approach by constructing a “cell-permeable CocE” via attachment of a thermostable variant of CocE to a cell penetrating peptide (CPP). The double mutant T172R/G173Q-CocE (dmCocE) with an in vitro half-life of 4.5 hours (~20-fold improvement over native CocE) has shown a robust antagonism against the toxicity and reinforcing effects of cocaine in animals6, 16, and therefore was chosen as the model for developing cell-permeable CocE in this paper. CPPs, also known as peptide transduction domains (PTDs) or membrane translocation sequences (MTSs), are a group of short (less than 30 amino acid) and positively-charged peptides that are able to cross the cell membrane. Aside from their own cellular uptake, via covalent or electrostatic linkage, CPPs can deliver a wide range of cell-impermeable cargos including therapeutic enzymes into the cell17. Although its detailed mechanism remains unclear, this CPP-mediated cellular uptake does not appear to cause any membrane perturbation or morphology changes of the cells11, 18. To date, CPPs have been successfully applied to create cell permeable forms of therapeutic enzymes in more than 300 studies, and this number is still growing 19.
Two model CPPs were chosen to develop the cell-permeable CocE variants in this study. The first one is Tat, a CPP derived from the HIV transcriptional trans-activator with the sequence YGRKKRRQRRR20. As the most intensively studied CPP, Tat has been employed in the clinical studies of several therapeutic macromolecules including a protein kinase C inhibitor and siRNA19, 21. Both in vitro and in vivo studies in Tat-mediated cellular uptake have demonstrated that Tat does not cause cell toxicity22 or increase the immunogenicity of its cargoes23. The second model CPP is the low molecular weight protamine (LWMP), which was developed in our laboratory and has the sequence VSRRRRRRGGRRRR24. LMWP possesses the cell-penetrating activity and kinetics comparable to those of Tat25, and has also been applied to deliver therapeutic macromolecules for the treatment of solid tumors, acute lymphoblastic leukemia, and hypoxic-angiogenesis26–28. In addition to its cell penetrating ability, LMWP offers several exclusive advantages. LMWP can be manufactured in mass quantities by a direct enzymatic digestion of protamine, which has been fully optimized into a single-step manufacturing process29. A further advantage is that, since protamine is an FDA-approved clinical drug, its derivative LMWP is also expected to be safe for humans. This proposition has been supported by the in vivo toxicity profile of LMWP, which demonstrates no acute toxicity30 or immunogenicity31, 32 in both rodent and canine models.
In this paper, we employed two different methods, chemical conjugation and genetic recombination, to produce CPP-attached dmCocE (CPP-dmCocE) variants. In the chemical conjugation method, CPP and dmCocE were linked via a disulfide bond that should be automatically cleaved in the reductive intracellular environment18. This self-detachable property of the CPP was designed to abort any potential interference with the conjugated dmCocE. In the genetic recombination method, the genes of the CPP and dmCocE were cloned and expressed in tandem, producing a recombinant CPP-dmCocE fusion protein. This strategy provides better control of the CPP number (e.g., one CPP per dmCocE molecule) and location (e.g., at the N- or C-terminus of dmCocE) on the fusion protein, since its primary structure is precisely encoded in the DNA sequence. The in vitro properties including the absolute yield per batch, recovery percentage, enzyme specific activities, and in vitro stability at 37 °C of these CPP-dmCocE variants were then systematically characterized and compared. The cellular uptake ability of all CPP-dmCocE variants were examined in HeLa cells, a human cervical carcinoma cell line that has been extensively employed in the studies of CPP-mediated intracellular delivery27, 33, 34. Our findings demonstrated that the cell-permeable and cocaine-hydrolyzing dmCocE could serve as not only a promising candidate for developing a non-disruptive cell-encapsulated CocE, but also a model in preparing other cell-impermeable therapeutic enzymes with strong immunogenicity that prevents them for possible clinical uses.
Materials and Experiments
Materials
pET22b(+)-dmCocE was employed to lead to an over expression of dmCocE with a C-terminal 6-histidine tag (LEHHHHHH) for purification35. Escherichia coli (E. coli) competent cells (DH5α and BL21 Star™ (DE3)), succinimidyl 3-(2-pyridyldithio) propionate (SPDP), pEXP-5-NT/TOPO® TA expression kit, minimum essential medium α (MEM-α), fetal bovine serum (FBS), 1X PBS (pH 7.4), Hoechst 33258, and Hank’s balanced salt solution (HBSS) were purchased from Invitrogen (Carlsbad, CA). Carbenicillin, isopropyl-β-thiogalactopyranoside (IPTG), dithiothreitol (DTT), leupeptin, soybean trypsin inhibitor, fluorescein isothiocyanate (FITC), and heparin sulfate were purchased from Sigma-Aldrich (St. Louis, MO). Tat peptide (H2N-YGRKKRRQRRR-COOH) was synthesized with approximately 98% purity by GenWay Biotech Inc. (San Diego, CA). LMWP was produced based on previously described procedures29. DNA primers used for site-directed mutagenesis and sequencing were synthesized by Integrated DNA Technologies Inc. (Coralville, IA). The Phusion® site-directed mutagenesis kit and DNA restriction endonucleases (NdeI, NcoI, KpnI, BamHI) were purchased from New England Biolabs (Ipswich, MA). Cocaine hydrochloride was obtained from the National Institutes on Drug Abuse (Bethesda, MA). The Bradford protein assay kit was purchased from Bio-Rad Laboratories (Hercules, CA). The human cervical carcinoma cell line HeLa was obtained from the American Type Culture Collection (Manassas, VA). Water was distilled and deionized. All other reagents were of analytical grade.
Expression and purification of dmCocE
dmCocE was produced according to procedures described previously6 with modifications. E. coli BL21 Star™ (DE3) cells were transformed with pET22b(+)-dmCocE plasmid, and then grown in 4-liter LB medium with 50 µg/mL of carbenicillin at 37°C. The expression of dmCocE was induced by the addition of 1 mM IPTG. After incubating for 12 hours at 18 °C, the BL21 Star™ (DE3) cells were harvested by centrifugation at 4000 × g and 37 °C for 30 min, and the cell pellets containing dmCocE were frozen at −80 °C overnight. The harvested cell pellets containing dmCocE were re-suspended in a purification buffer containing 50 mM Tris-HCl and 150 mM NaCl at pH 8.0, and then lysed with lysozyme (0.5 mg/mL) and DNase (8 µg/mL) in the presence of 1mM MgCl2. The histidine-tagged dmCocE was purified by Talon™ metal affinity chromatography (Clontech Laboratories Inc., Mountain View, CA) and Q-Sepharose anion-exchange chromatography (GE Healthcare, Piscataway, NJ). dmCocE was eluted from the Q-Sepharose column with a 250–450 mM NaCl linear gradient (pH 7.4) containing 20 mM HEPES, 2 mM MgCl2, 1 mM EDTA, and 1 mM DTT. The peak fractions were pooled and concentrated using an ultra-filtration device (molecular weight cut-off: 30,000 Da), flash frozen in liquid nitrogen, and then stored at −80 °C prior to use. All purification procedures were carried out at 4 °C in presence of protease inhibitors (1 µM leupeptin and 5 µg/mL soybean trypsin inhibitor).
Preparation of CPP-S-S-dmCocE chemical conjugates
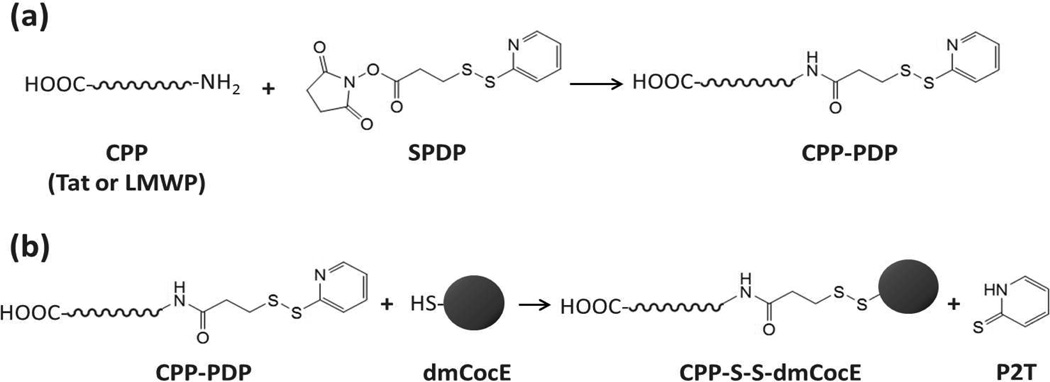
In the chemical conjugation approach, the scheme utilizing the sulfhydryl groups of dmCocE was selected (Figure 1), because any modification to the primary amine groups of dmCocE led to a loss of its enzymatic activity8. Before undergoing conjugation, CPP was modified to introduce an N-terminal reactive sulfhydryl group using a bifunctional cross-linker SPDP (Figure 1a). Five molar excess of SPDP in dimethyl sulfoxide (DMSO) was mixed with Tat or LMWP solution (5 mg/mL in a 0.1 M phosphate buffer, pH 8.0), and incubated at room temperature for 2 hours with gentle shaking. The sulfhydryl contents of SPDP-modified Tat and LWMP, namely PDP-Tat and PDP-LMWP, were measured by pyridine-2-thione assay36. In a separate process, dmCocE was pre-treated with DTT (10 mM) at 4 °C for 2 hours to fully restore its surface sulfhydryl groups. The DTT in dmCocE solution was then removed using an Amicon Ultra-15 centrifugal filter device (molecular weight cut-off: 30,000 Da).
Figure 1.
Scheme of chemical conjugation of CPP to dmCocE by using SPDP as the crosslinker.
The conjugation reaction was performed by mixing the DTT-pretreated dmCocE solution with three molar excess of PDP-Tat or PDP-LMWP (Figure 1b) and incubating at 4 °C for 12 hours. Unreacted PDP-Tat or PDP-LMWP was removed by size-exclusion chromatography using a Sephadex G-25 desalting column (GE Healthcare, Piscataway NJ). The CPP-conjugated dmCocE variants, namely Tat-S-S-dmCocE and LMWP-S-S-dmCocE, were separated from unreacted dmCocE by heparin affinity chromatography using a HiTrap heparin column (GE Healthcare, Piscataway NJ). Tat-S-S-dmCocE and LMWP-S-S-dmCocE were eluted from the heparin column with a linear NaCl gradient (pH 7.4) containing 20 mM HEPES, 2 mM MgCl2, and 1 mM EDTA. The two isolated CPP-S-S-dmCocE variants were then concentrated, flash frozen in liquid nitrogen, and stored at −80 °C prior to use.
Construction and preparation of recombinant CPP-dmCocE fusion proteins
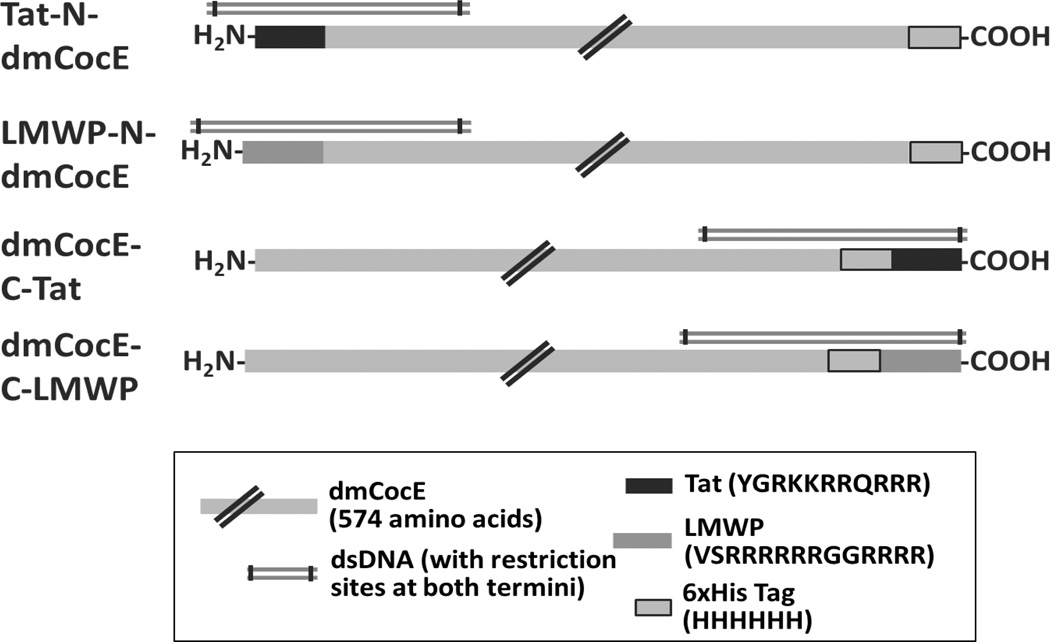
In the genetic recombination approach, four types of CPP-dmCocE fusion proteins were created by appending the DNA sequence encoding the CPP at the beginning or the end of dmCocE genes (Figure 2). To facilitate the insertion of a CPP sequence, a BamHI restriction site was introduced at the end of the C-terminal 6-histidine tag on pET22b(+)-dmCocE plasmid. A Phusion® site-directed mutagenesis protocol (New England Biolabs) with modifications37 was employed using one synthesized mutagenic primer: 5’-CCACCACCACCACTagGATCCGGCTGCTAACAAAGC-3’ (mutated codon showed in lowercase and BamHI site underlined). The incorporation of mutation was confirmed by the results of BamHI digestion and DNA sequencing (the DNA sequencing Core, University of Michigan).
Figure 2.
Schematic design for the four recombinant CPP-dmCocE fusion proteins. Double-stranded DNA fragments (dsDNA) with restriction sites at both termini were generated by a mutagenic PCR to facilitate the insertion of CPP sequences; see the Materials and Experiments section for detailed information.
Four double-stranded DNA fragments containing the first or the last ~450 bases of dmCocE and CPP (Tat or LMWP) codons were produced by a mutagenic PCR to append CPP (Tat or LMWP) sequence to dmCocE gene. The sequences of synthesized CPP-tailed primers are available in the Supporting Information, which is available free of charge via the Internet at http://pubs.acs.org. For better specificity and higher success rate of sub-cloning, the four PCR products were first inserted to pEXP-5-NT/TOPO® vectors using the vendor’s protocol (Invitrogen), and then sub-cloned to the mutated pET22b(+)-dmCocE plasmid, as described previously. During sub-cloning procedures, both the mutated pET22b(+)-dmCocE plasmid and the four DNA inserts (on pEXP-5-NT/TOPO® vectors) were double digested with either NdeI/NcoI or KpnI/BamHI. NdeI and NcoI generated overhangs at the beginning, while KpnI and BamHI generated overhangs at the end of the dmCocE gene. The digested plasmid and DNA inserts were then purified by 1% agarose gel electrophoresis, ligated by T4 ligase (16 °C, 18 hours), and transformed into DH5α competent cells. The cells were grown overnight at 37 °C on agar plates containing 50 mg/mL carbenicillin. Plasmids from individual colonies were isolated, and the recombinant CPP-dmCocE genes were confirmed by sequencing of both strands over the entire coding region (the DNA sequencing Core, University of Michigan). The plasmids containing the CPP-dmCocE genes were then re-transformed into E. coli BL21 Star™ (DE3) cells for the expression of recombinant CPP-dmCocEs. The expression and purification procedures used for preparing dmCocE were also utilized to produce CPP-dmCocE fusion proteins, and therefore not repeated here.
Protein assays
At each purification step, the protein contents and purity were examined using SDS-PAGE. Gels were stained with coomassie blue, and the images were analyzed with ImageJ (National Institutes of Health, Bethesda, MD). Protein concentration was determined by the Bradford protein assay kit using bovine serum albumin (BSA) as standard. Each sample was done in triplicate with at least five independent dilutions (n ≥ 5).
Cocaine hydrolysis kinetic analysis
The cocaine hydrolyzing activities of the CPP-dmCocE constructs were determined by a real-time spectrophotometric assay developed by Larsen et al.38 with modifications. Cocaine hydrolyzing reactions were conducted in a UV-transparent 96-well microplate, and initiated by adding 100 µL of CPP-dmCocE solutions to 100 µL cocaine solutions. The final concentration of CPP-dmCocE solutions was 50 ng/mL. Final cocaine concentrations were 1, 2.5, 5, 10, 25, 50, 100, and 250 µM. CPP-dmCocE and cocaine solutions were prepared in phosphate-buffered saline (PBS) at pH 7.4 (137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate dibasic, and 2 mM potassium phosphate monobasic, pH of 7.4). To obtain steady-state kinetic parameters for CPP-dmCocE variants, the initial rate of cocaine hydrolysis (in micromole per minute) was determined by monitoring the decrease in absorbance at 240 nm, where the difference in molar absorptivity between cocaine and its metabolites (ecgonine methyl ester and benzoic acid) was 6,700 M−1 cm−1 39. Absorbance at 240 nm was monitored every 15 seconds throughout the 30-min time course using a PowerWave XS microplate spectrophotometer (Biotek Instruments, Inc., Winooski, VT). For the in vitro stability analysis, CPP-dmCocE constructs (100 ng/mL) in PBS (pH 7.4) were pre-incubated at 37 °C, and aliquots were taken at different time points (0.5, 1, 2, 3, 4, 6, 8, 10, and 12 hours). The remaining cocaine-hydrolyzing activity of the aliquots was assayed as described previously. All studies were done in triplicate with at least three independent measurements (n ≥ 3).
Data analysis and curve-fitting were conducted using Prism 5 (GraphPad Software, San Diego CA). For the cocaine hydrolyzing activity assays, the data points were fit via non-linear regression to the Michaelis-Menten equation, taking into account the enzyme concentrations. For the in vitro stability assays, the ratios of apparent of each sample aliquot, which were determined by the Michaelis-Menten kinetics and reflected the apparent cocaine-hydrolyzing efficiency, were fit to the single-phase exponential decay with pre-incubation time at 37 °C.
Cell culture
CPP-dmCocE constructs and dmCocE were labeled with FITC at their primary amino groups. In brief, CPP-dmCocE solutions (10 mg/mL in 0.1 M carbonate buffer, pH 9.2) were incubated in a 1:20 molar ratio with a FITC solution in dimethylformamide (DMF) at 4 °C overnight with gentle shaking. The final DMF concentration in the reaction mixture was less than 5%. After incubation, the excess FITC was removed using a Sephadex G-25 desalting column (GE Healthcare). The concentrations and degrees of labeling of the FITC-labeled dmCocE and CPP-CocE constructs were determined by measuring the absorbances at 280 nm and 494 nm. The FITC-labeled enzymes were then concentrated, flash frozen in liquid nitrogen, and stored at −80 °C prior to use.
HeLa cells were seeded at a density of 3 × 104 cells/cm2 in the BD PureCoat™ amine-surface 24-well plate (Bedford, MA), and grown in MEM-α medium supplemented with 10 % (v/v) FBS. After they reached a confluence at ~60 % (approximately 24 hours after seeding), cells were incubated with FITC-labeled enzymes (5 µM) in a FBS-free MEM-α medium at 37 °C for 2 hours. After incubation, the treated HeLa cells were washed three times with 10 mg/mL of heparin sulfate in PBS (pH 7.4). Heparin is added in the wash buffer to facilitate the removal of the non-internalized CPP-CocE 40, which is achieved by competing for binding of the heparan sulfate proteoglycans to the CPP moieties of the CPP-CocE products. The cell nuclei were counter-stained with 5 µg/mL of Hoechst 33258 in PBS for 20 min at 37 °C. To visually detect the cellular uptake, cell fluorescence was examined with a Nikon Eclipse TE2000S inverted fluorescence microscope using a 20X objective and three channels (DIC (differential interference contrast), Hoechst 33258, and FITC). Cell images were acquired and analyzed with Metamorph® software (Molecular Devices Corporation, Sunnyvale, CA).
Results
CPP-S-S-dmCocE chemical conjugates
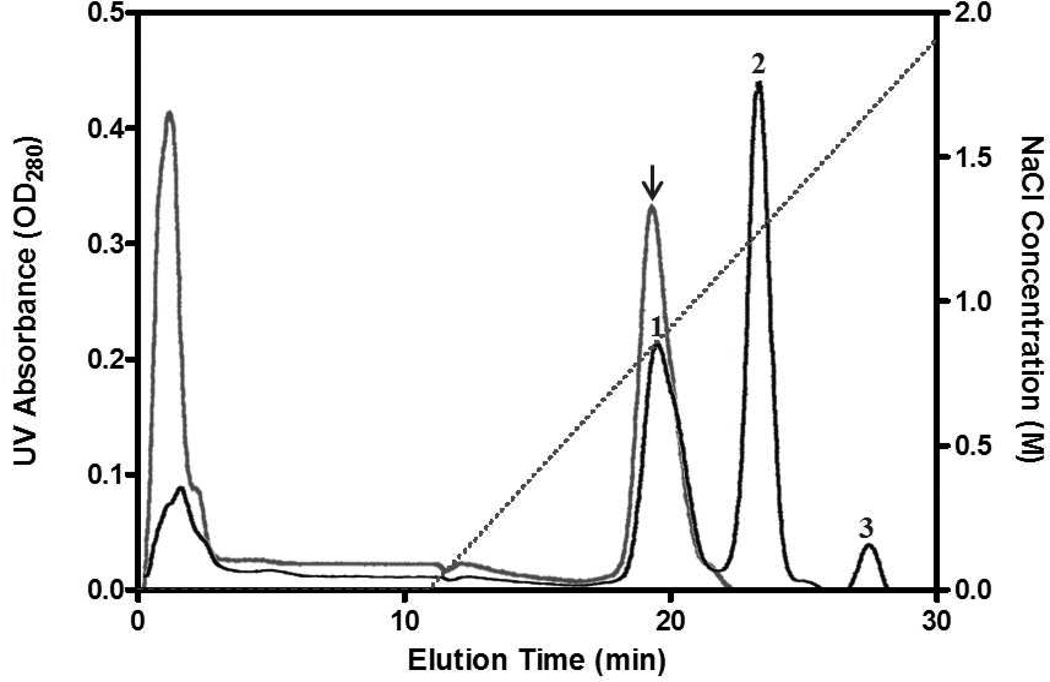
Two disulfide-bridged chemical conjugates, Tat-S-S-dmCocE and LMWP-S-S-dmCocE, were synthesized using a bi-functional cross-linker SPDP. The elution profile (Figure 3) indicated that both of these chemical CPP-dmCocE conjugates could be purified by heparin affinity chromatography. In the absence of a CPP group, the unreacted dmCocE displayed no binding affinity to the heparin column, and eluted before the salt gradient started. In contrast, Tat-S-S-dmCocE and LWMP-S-S-dmCocE bound to the heparin column and eluted at ~0.8 M or higher concentrations of NaCl, indicating that these two CPP-CocE chemical conjugates bind strongly to the heparin column.
Figure 3.
Chromatograms of Tat-S-S-dmCocE (gray line) and LMWP-S-S-dmCocE (black line) from a heparin column. The NaCl concentration applied in elution was shown in dotted line. Tat-S-S-dmCocE (indicated in arrow) eluted at 0.75 M NaCl whereas LMWP-S-S-dmCocE eluted in three peaks at 0.8 M (Peak #1), 1.2 M (Peak #2), and 1.6 M (Peak #3) of NaCl, respectively. Unreacted dmCocE eluted at the beginning without NaCl gradient.
Another interesting observation from the heparin chromatogram is that, during the chemical conjugation reaction between LMWP-PDP and dmCocE, at least three variants of LWMP-S-S-dmCocE, likely with different numbers and/or locations of LMWP, were generated. Based on its backbone and three-dimensional structure, each CocE (and dmCocE) molecule has four sulfhydryl groups, three of which are on the surface and accessible to chemical conjugation38. Moreover, LMWP itself is known to have a strong binding affinity to a heparin column and can only elute at 1.0 M or higher concentrations of NaCl29. Thus, it is reasonable to suggest that these three LMWP-S-S-dmCocE variants eluting at 0.8, 1.2, and 1.6 M NaCl have one, two, and three LMWP groups conjugated on each dmCocE molecule. On the other hand, only one variant of Tat-S-S-dmCocE was formed during the chemical conjugation reaction between Tat-PDP and dmCocE. The ratio of unreacted dmCocE in this conjugation reaction was also found to be higher than that in the LMWP-S-S-dmCocE conjugation reaction.
Table 1 shows the yield and activity of CPP-S-S-dmCocE chemical conjugates. The amount of chemical conjugate produced from 16 mg dmCocE is 3.9 mg in LMWP-S-S-dmCocE, and 1.05 mg in Tat-S-S-dmCocE. For LMWP-S-S-dmCocE, that amount equals 18.5% of yield in activity. The yield in activity of Tat-S-S-dmCocE was not determined, because the amount generated was only sufficient for the subsequent cellular uptake studies.
Table 1.
Preparation of CPP-S-S-dmCocE chemical conjugates.
| Total Proteina | Specific Activitya | Total Activity |
Yieldb | ||
|---|---|---|---|---|---|
| (mg) | (U/mg) | (U) | (%) | ||
| dmCocE (Reactant) | 15.83 (±1.13) | 41.4 (±1.2) | 655 | 100 | |
| Tat-S-S-dmCocE | 1.05 (±0.06) | Not Determinedd | |||
| LMWP-S-S-dmCocE | #1c | 1.13 (±0.06) | 36.7 (±3.3) | 41 | 18.5 |
| #2c | 2.10 (±0.09) | 28.0 (±0.9) | 59 | ||
| #3c | 0.67 (±0.07) | 31.0 (±1.2) | 21 | ||
Standard errors present in parentheses.
Compared with the total activity of dmCocE.
Refers to the three peaks of LMWP-S-S-dmCocE isolated by a heparin affinity column (see Figure 3).
Due to the lack of a sufficient amount of the current product for analysis.
Recombinant CPP-dmCocE Fusion Proteins
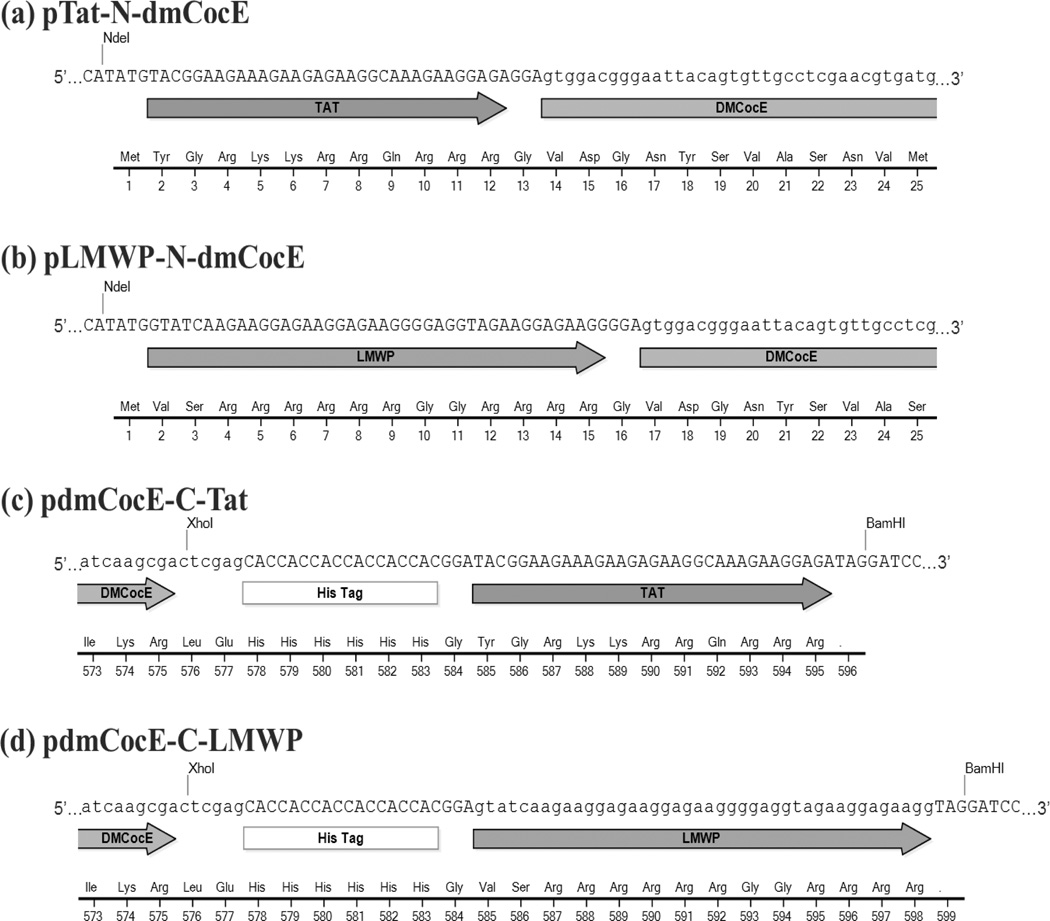
Four recombinant vectors derived from pET22b(+)-dmCocE were constructed to produce CPP-dmCocE fusion proteins in E. coli. pTat-N-dmCocE (Figure 4a) and pLMWP-N-dmCocE (Figure 4b) both have a CPP sequence appended at the beginning of dmCocE gene. In contrast, pdmCocE-C-Tat (Figure 4c) and pdmCocE-C-LMWP (Figure 4d) both have a CPP sequence at the end of the C-terminal 6-histidine tag on the dmCocE gene. An additional glycine (Gly) residue was inserted between CPP and dmCocE (for pTat-N-dmCocE and pLWMP-N-dmCocE) or the C-terminal 6-histidine tag (for pdmCocE-C-Tat and pdmCocE-C-LMWP) as a spacer to allow for greater flexibility of the CPP chains.
Figure 4.
Partial schematic maps of DNA/protein sequence in the beginning (a, b) or the end (c, d) regions of recombinant CPP-dmCocE fusion proteins on the corresponding expression vectors. Except BamHI sites, all other restriction sites (NdeI, XhoI) are from the original pET22b(+)-dmCocE vector.
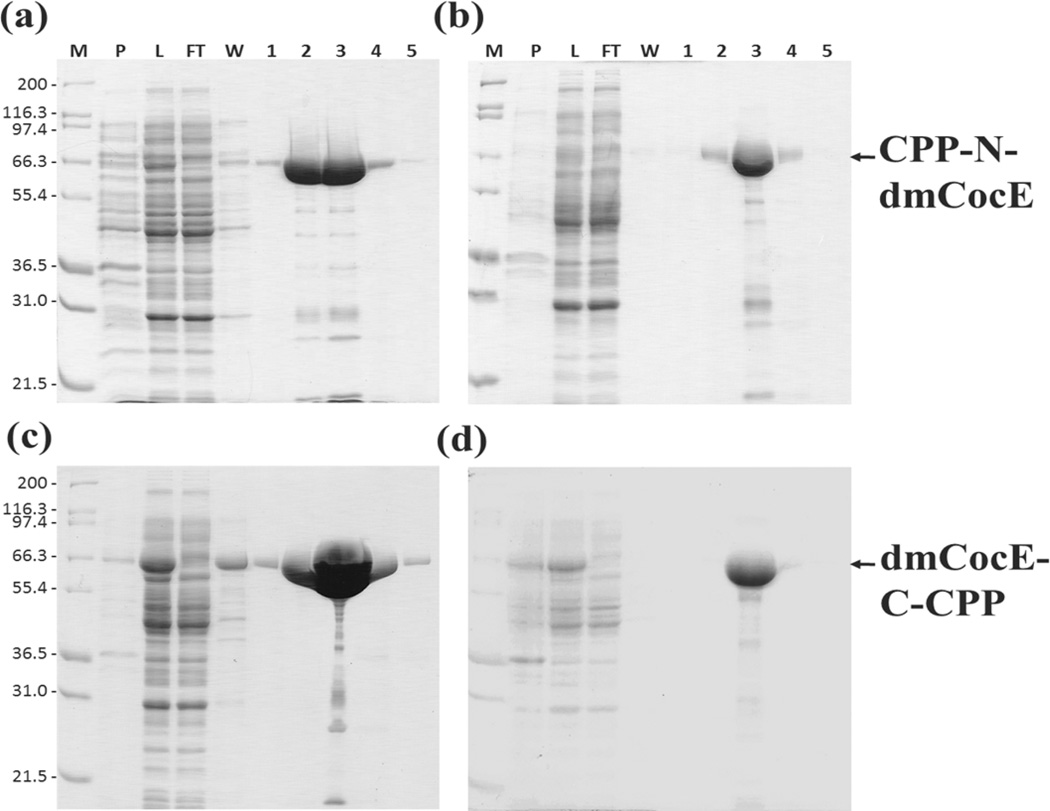
These four CPP-dmCocE fusion proteins were expressed as soluble, histidine-tagged proteins in E. coli; only small amounts were insoluble and found in the pellet of cell lysate (Figure 5). Very little of the CPP-dmCocE fusion protein appeared in the flow-through or in the wash buffer containing 5 µM imidazole, indicating that the binding affinity between the 6-histidine tag and the Co2+ metal ion in the Talon™ column was not impaired by the addition of the CPP groups. Contaminants in the crude eluate of the four CPP-dmCocE fusion proteins from a Talon™ column were further separated by Q-Sepharose anion-exchange chromatography. After Q-sepharose purification, the purity of the CPP-dmCocE products was at least 99%, which were determined by image analysis of SDS-PAGE results.
Figure 5.
SDS-PAGE analysis of expression and purification of four CPP-dmCocE fusion proteins (a) Tat-N-dmCocE, (b) LMWP-N-dmCocE, (c) dmCocE-C-Tat, and (d) dmCocE-C-LMWP. Lane M: Mark12™ protein molecular weight standard (Invitrogen); Lane P: the pellet fraction of cell lysate; Lane L: the supernatant fraction of cell lysate; Lane FT: flow-through fraction; Lane W: wash fraction; Lane 1–5: elution fraction containing dmCocE. Lane FT to Lane 5 were obtained after passing through from the Talon™ column.
The recovery of four CPP-dmCocE fusion proteins in each purification step is shown in Table 2. Approximately 700 mg of total soluble protein was produced from the cultures expressing CPP-dmCocE fusion proteins, which was similar to the expression of native dmCocE in E. coli (data not shown). The first purification step, Co2+ metal affinity chromatography, successfully enriched the CPP-dmCocE fusion proteins in the cell lysate supernatant. From the initial cell lysate supernatant to the Talon™ column eluate, the specific activity of cocaine hydrolysis increased from less than 5 U/mg to approximately 30 U/mg, indicating an at least six-fold increase. However, a 20–30% protein loss also occurred after the Co2+ metal affinity chromatography. This protein loss may be attributed to the relatively small amount of protein eluted in flow-through and wash buffer, based on the analysis of the SDS-PAGE results (see Figure 5). The second purification step, Q-Sepharose anion-exchange chromatography, successfully separated the impurities co-eluting with the CPP-dmCocE fusion proteins from the Talon™ column. According to Table 2, the recovery from the Talon™ column purification to the Q-sepharose chromatography was at least 97%, and the specific enzyme activity was further increased to around 30 U/mg. This consistent specific activity among the four fusion proteins implied that the purification scheme works consistently, and the percentage yield after purification was expected to be higher because of the higher initial mass quantity. Regarding the final yields in mass quantity, the CPP-dmCocE produced in the highest mass quantity (mg) was dmCocE-C-Tat (73 mg), followed by Tat-N-dmCocE (33 mg), dmCocE-C-LMWP (10 mg), and then LMWP-N-dmCocE (7 mg). In general, the amount of CPP-dmCocE fusion proteins accounts for 1–10% of the total soluble proteins in E. coli, and the C-terminal, Tat-attached CPP-dmCocE variants showed higher production yield compared with the N-terminal, LMWP-attached variants.
Table 2.
Step-wise purification of recombinant CPP-dmCocE fusion proteins from the culture supernatant of E. coli.
| CPP-dmCocE Fusion Proteins |
Fraction | Total Protein |
Specific Activity |
Total Activity |
Yield |
|---|---|---|---|---|---|
| (mg) | (U/mg) | (U) | % | ||
| Tat-N-dmCocE | Cell Lysate Supernatant | 720.6 (±29.0) |
2.0 (±0.2) |
1421.3 | 100 |
| Co-Sepharose CL (Talon) |
34.8 (±0.9) |
28.9 (±1.1) |
1006.1 | 70.8 | |
| Q-Sepharose HP | 32.7 (±0.2) |
30.1 (±0.6) |
985.9 | 69.4 | |
| LMWP-N-dmCocE | Cell Lysate Supernatant | 758.7 (±32.4) |
0.4 (±0.2) |
286.3 | 100 |
| Co-Sepharose CL (Talon) |
8.0 (±0.1) |
29.9 (±0.3) |
239.4 | 83.6 | |
| Q-Sepharose HP | 7.3 (±0.1) |
31.8 (±0.5) |
231.9 | 81.0 | |
| dmCocE-C-Tat | Cell Lysate Supernatant | 727.9 (±39.6) |
4.8 (±0.3) |
3494.5 | 100 |
| Co-Sepharose CL (Talon) |
75.8 (±2.5) |
31.6 (±1.1) |
2398.9 | 68.6 | |
| Q-Sepharose HP | 73.3 (±2.3) |
32.5 (±0.6) |
2379.4 | 68.1 | |
| dmCocE-C-LMWP | Cell Lysate Supernatant | 704.1 (±32.6) |
0.5 (±0.1) |
371.5 | 100 |
| Co-Sepharose CL (Talon) |
10.6 (±0.3) |
28.3 (±0.6) |
300.4 | 80.9 | |
| Q-Sepharose HP | 10.3 (±0.3) |
29.0 (±0.7) |
299.3 | 80.6 |
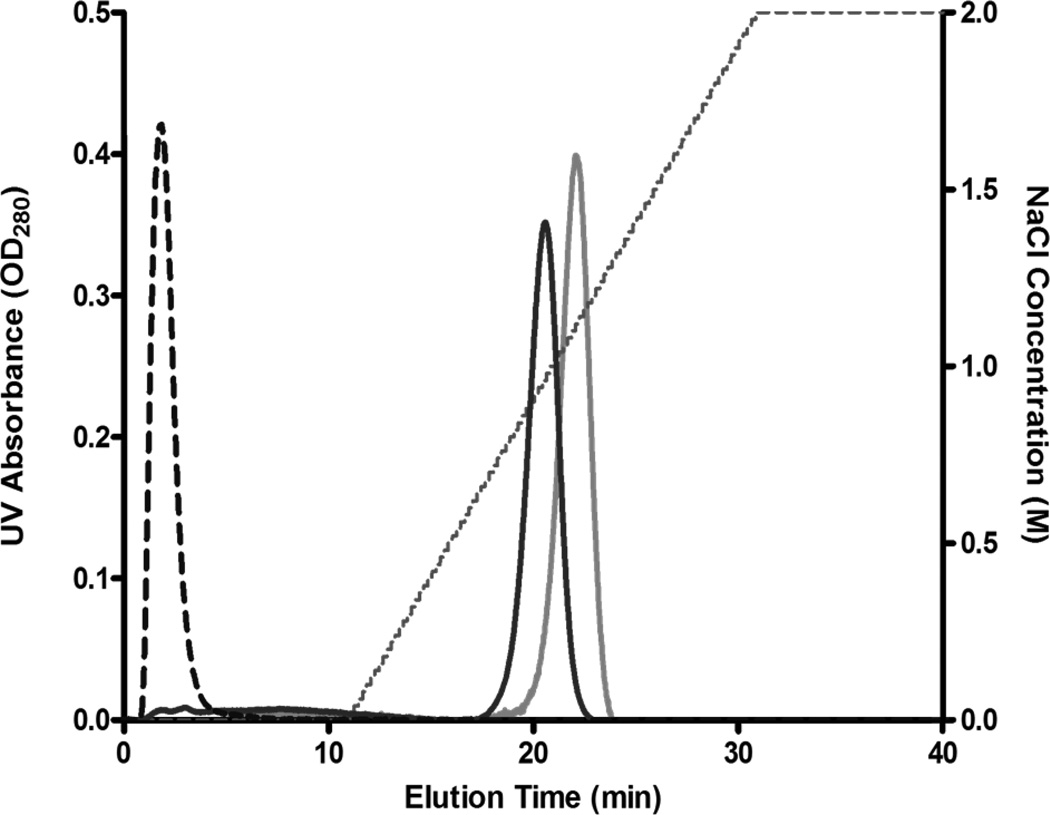
Since chemical CPP-dmCocE conjugates have shown a strong binding affinity to the heparin column (see Figure 3), it was of interest to determine if recombinant CPP-dmCocE fusion proteins also possess a similar binding affinity. The elution chromatogram of Tat-N-dmCocE and dmCocE-C-Tat (Figure 6) indicated that Tat-dmCocE products were also able to bind to a heparin affinity column, and did not elute until approximately 1 M NaCl. In contrast, native dmCocE showed no binding affinity to the heparin column and eluted before the salt gradient. This heparin binding affinity of CPP-dmCocE fusion proteins (and chemical CPP-dmCocE conjugates) to a heparin column indicates that the CPP groups were exposed on the enzyme surface, which was favorable for the interaction with the negatively charged extracellular proteins and hence should facilitate cellular uptake.
Figure 6.
Chromatograms of Tat-N-dmCocE (black line) and dmCocE-C-Tat (gray line) from a heparin column. The NaCl concentration applied in elution is shown in dotted line. Tat-N-dmCocE eluted at 0.9 M NaCl, whereas dmCocE-C-Tat eluted at 1.2 M NaCl. As a reference, dmCocE (dashed line) eluted at the beginning without NaCl gradient.
Cocaine Hydrolyzing Activity
The specific cocaine hydrolyzing activities of the disulfide-bridged LMWP-S-S-dmCocE chemical conjugates, as well as the four CPP-dmCocE fusion proteins, are shown in Table 3. The three peaks of LMWP-S-S-dmCocE chemical conjugates seen in Figure 3 have cocaine-hydrolyzing efficiency slightly lower than that of native dmCocE. On the other hand, compared with native dmCocE, the four fusion proteins show a ~50% reduction of cocaine-hydrolyzing efficiency. This reduction in cocaine-hydrolyzing activity apparently came from their slightly lower kcat, and significantly higher KM (ranging from 30% to 70% of increase) relative to native dmCocE. Among these fusion proteins, Tat-N-dmCocE has the lowest KM, and therefore possesses the highest affinity towards cocaine as well as the highest cocaine-hydrolyzing efficiency of .
Table 3.
Kinetic parameters of dmCocE and CPP-dmCocE constructs in hydrolyzing cocaine.
| dmCocE Variants | kcat | KM | kcat/KMa | |||
|---|---|---|---|---|---|---|
| (min−1) | (µM) | (min−1µM−1) | ||||
| Original dmCocE | 2691 (±77) | 28.8 (±2.6) | 93.6 (±6.4) | |||
| LMWP-S-S-dmCocE | #1 | 2350 (±218) | 30.5 (±8.8) | 77.0 (±16.7) | ||
| #2 | 1795 (±61) | 21.5 (±2.5) | 83.7 (±7.6) | |||
| #3 | 1975 (±87) | 23.1 (±3.4) | 85.6(±9.8) | |||
|
CPP-dmCocE Fusion Proteins |
Tat-N-dmCocE | 1957 (±38) | 39.6 (±2.2) | 49.4 (±2.0) | ||
| LMWP-N-dmCocE | 2065 (±32) | 49.0 (±2.0) | 42.1 (±1.2) | |||
| dmCocE-C-Tat | 1887 (±41) | 30.6 (±2.1) | 61.7 (±3.2) | |||
| dmCocE-C-LMWP | 2110 (±45) | 41.5 (±2.5) | 50.8 (±2.2) | |||
Derived from fit to the equation .
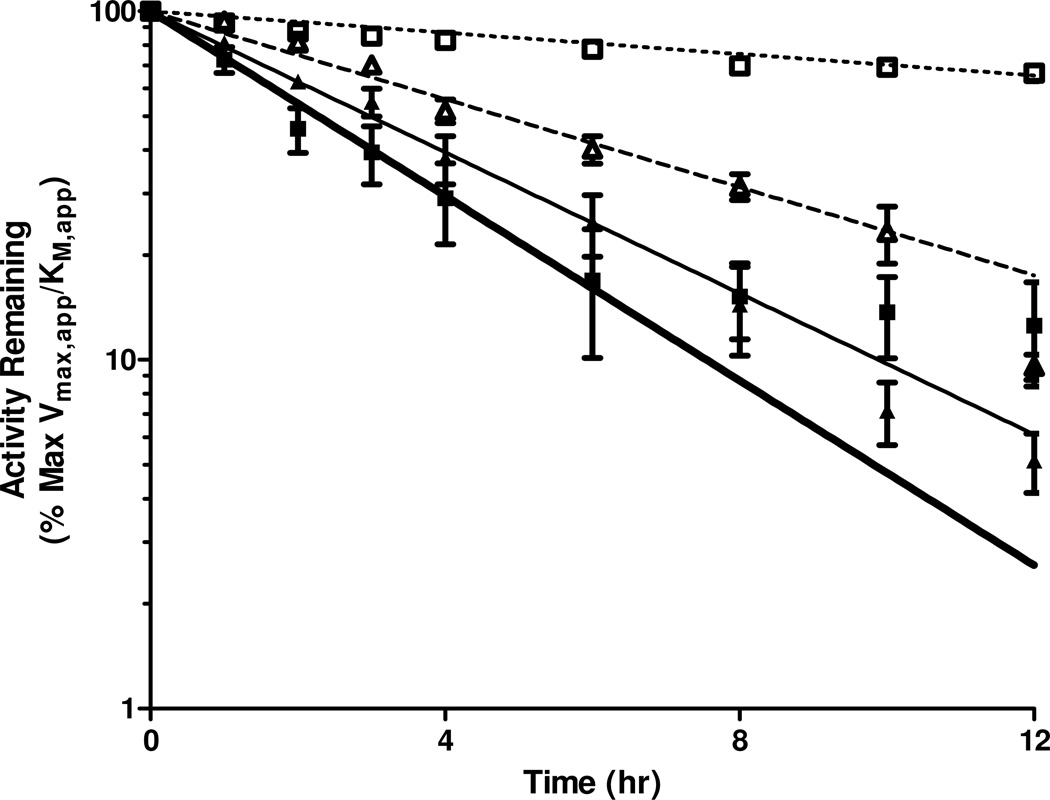
In vitro Stability at 37 °C
The in vitro stabilities at 37 °C of the four CPP-dmCocE fusion proteins were assessed by monitoring their apparent cocaine-hydrolyzing efficiency () over time (Table 4, Figure 7). The attachment of the CPP groups had different impacts on the stability of each CPP-CocE fusion protein. Compared with native dmCocE, the two C-terminal CPP-dmCocE variants (Tat-C-dmCocE and LMWP-C-dmCocE) were less stable, whereas LMWP-N-dmCocE possessed a half-life and MRT (mean residence time; the average duration that a dmCocE or CPP-dmCocE molecule remains active) similar to those of dmCocE. Interestingly, Tat-N-dmCocE displayed a prolonged half-life and MRT; which were 1.3-fold higher over that of dmCocE, and was comparable to the most stable CocE variant reported in the literature7. Based on an additional in vitro stability study containing prolonged time points, Tat-N-dmCocE still maintained 52% of its cocaine-hydrolyzing efficiency even after incubation at 37 °C for 24 hours (data not shown). Another interesting observation is that the efficiency decay curve of dmCocE-C-Tat (Figure 7, solid squares) did not completely follow the anticipated one-phase exponential decay during the 12-hour incubation (Figure 7, broad solid line) but showed a slower decrease after incubating for six hours. This observation did not occur in the other three CPP-dmCocE fusion proteins, native CocE, or any known CocE variants reported previously4, 6, 7.
Table 4.
In vitro stability kinetics of dmCocE and CPP-dmCocE fusion proteins following a 12-hour incubation at 37 °C.
| CocE Variants | kcat,app | KM,app | t1/2 | MRTa | ||
|---|---|---|---|---|---|---|
| (min−1) | (µM) | (min−1µM−1) | (hr) | (hr) | ||
| dmCocE | 4.28b | 6.17b | ||||
| Tat-N-dmCocE | 1972 (±71) | 69.8 (±6.2) | 28.2 (±1.6) | 5.76 (±1.45) | 8.31 (±2.09) | |
| LMWP-N-dmCocE | 651 (±59) | 159.2 (±27.5) | 4.1 (±0.4) | 4.83 (±1.78) | 6.96 (±2.57) | |
| dmCocE-C-Tat | 707 (±81) | 114.2 (±27.8) | 6.2 (±0.9) | 1.66 (±0.39) | 2.39 (±0.56) | |
| dmCocE-C-LMWP | 1043 (±273) | 518.0 (±186.5) | 2.0 (±0.2) | 2.98 (±0.57) | 4.29 (±0.83) |
Mean residence time.
From Narasimhan et al.35; standard errors not available.
Figure 7.
In vitro thermal stability of four CPP-dmCocE fusion proteins at 37 C°. 100 ng/mL of Tat-N-dmCocE (open square), LMWP-N-dmCocE (open triangle), dmCocE-C-Tat (solid square), and dmCocE-C-LMWP (solid triangle) in PBS (pH 7.4) were pre-incubated at 37 °C, and aliquots were taken at different time points. The remaining cocaine-hydrolyzing efficiency of the aliquots, which was represented by the ratio of apparent kcat over KM (), was evaluated as percentage of the value of the non-incubated CPP-dmCocE fusion proteins, and was then fit to the single-phase exponential decay with pre-incubation time.
Cellular Uptake behavior of Six CPP-dmCocE Variants
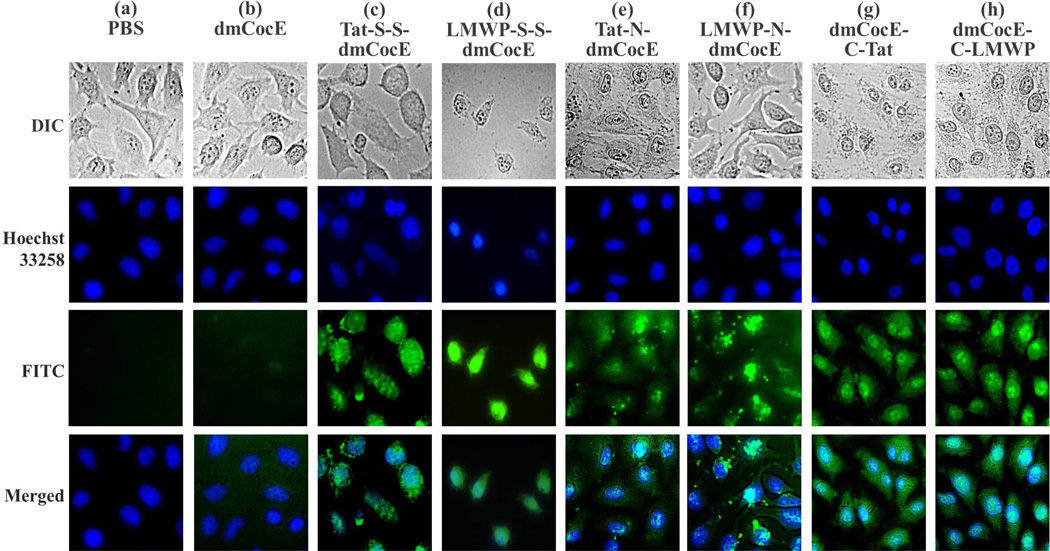
After incubating with HeLa cells for 2 hours at 37 °C, at a concentration of 5 µM, all six FITC-labeled CPP-dmCocE constructs penetrated and accumulated into the living HeLa cells, generating a significant intracellular fluorescence (Figure 8). In comparison, FITC-labeled dmCocE at the same concentration showed no cell penetrating activity, and its fluorescence signals were almost non-detectable after wash. Hence, the cell penetrating abilities of the CPP-dmCocE constructs were obviously attributed to the attached CPP groups, namely Tat or LMWP. In addition, the morphologies of the cells treated with CPP-dmCocE appeared normal and similar to those of PBS-treated cells. The normal cell morphology of the CPP-dmCocE treated cells excluded the possibility that the uptake was due to cell membrane disruption. This conclusion was further supported by trypan blue exclusion, which indicated the membrane integrity of the cells loaded with CPP-dmCocE variants was not compromised (data not shown).
Figure 8.
Internalization of six CPP-dmCocE constructs in HeLa cells. The cells were incubated with 5 µM of dmCocE (b) or CPP-dmCocE constructs (c–h) for 2 hours at 37 °C. After incubation, cells were washed extensively with 10 mg/mL heparin in PBS, counterstained the nuclei with Hoechst 33258, and their images were then acquired by fluorescence microscopy. Nucleus was detected from the Hoechst 33258 channel (blue), and dmCocE and CPP-dmCocEs were detected from the FITC channel (green). Cell morphology was acquired from the DIC microscopy in gray scale.
Although all six CPP-dmCocE constructs possessed cell-penetrating activity, their distribution after cell internalization showed different phenotypes. First, compared with the CPP-dmCocE chemical conjugates, CPP-dmCocE fusion proteins exhibited a clear enrichment in the nucleus (Figure 8e–h). This nuclear accumulation of CPP-dmCocE fusion proteins was even more apparent for dmCocE-C-Tat and dmCocE-C-LMWP (Figure 8g and 8h). Since Tat and LMWP are also known to have DNA-binding activity 41, 42, the nuclear accumulation of these four CPP-dmCocE fusion proteins may be accounted for in terms of the ability to cross the nuclear membrane and bind to DNA. Second, some CPP-dmCocE constructs showed a homogenous distribution in the cytoplasm. The chemical conjugate LMWP-S-S-dmCocE (Figure 8d), as well as two fusion proteins dmCocE-C-Tat (Figure 8g) and dmCocE-C-LMWP (Figure 8h), dispersed evenly and without any condensation in the cytoplasm. This homogenous distribution suggests that after cell internalization these three CPP-dmCocE constructs were not confined in the cytoplasmic sub-compartments such as endosomes. In comparison, the other chemical conjugate (Tat-S-S-dmCocE, Figure 8c) and fusion proteins (N-Tat-dmCocE and N-LMWP-dmCocE, Figure 8e and 8f) showed a “spotty” distribution in the cytoplasm. In general, the two chemical conjugates showed primarily cytoplasmic distribution, while all the four recombinant fusion proteins displayed both nuclear and cytoplasmic localization, with dmCocE-C-CPP exhibiting higher cytoplasmic distribution during cellular uptake.
Discussion
In this research, we developed CPP-dmCocE variants by linking CPP and dmCocE via either chemical conjugation or genetic recombination. Each of these two strategies possesses its own advantages. The chemical conjugation strategy potentially presents a major advantage owing to the formation of cleavable disulfide linkages. Once the disulfide-bridged CPP-dmCocE conjugates enter the reductive intracellular environment, the linkage between the CPP and dmCocE should be rapidly cleaved, resulting in the release of dmCocE. This self-detachable behavior can minimize the potential interference from CPP, and therefore maintain the full enzymatic activity of dmCocE. More importantly, without the coupled CPP group, dmCocE itself is not cell permeable and will be permanently entrapped inside the cytoplasm. Thus, CPP detachment should prevent any potential backward translocation of dmCocE from the cytoplasm to the extracellular environment, and guarantee the effectiveness of cell encapsulation.
Despite the advantage of having a detachable CPP, these CPP-dmCocE chemical conjugates also possess two major drawbacks. First, since an activated CPP could randomly attach to any of the three surface sulfhydryl groups on dmCocE, the location and number of CPP on each chemical conjugate would vary. Indeed, the elution profile from the heparin affinity chromatography (Figure 3) is consistent with three forms of LWMP-S-S-dmCocE with different number and/or location of LMWP in this preparation. Second, since not all of the reactants (CPP and dmCocE) could be converted into desired products (CPP-dmCocE conjugates), an additional purification step was required to separate the conjugates from unreacted substrates, and the final yields of both chemical conjugations in mass quantity were low (25% and 7%; see Table 1). The yield of chemical conjugation could be scaled up; however, to achieve a yield similar to those of CPP-dmCocE fusion proteins, the expense of a scaled-up conjugation approach would be ~1000 times higher than that of a single-step genetic recombination approach, based on a similar laboratory setup as used in this study. The aforementioned two limitations were consistent with our observations, and would potentially occur to all of the CPP-protein products derived from the chemical conjugation strategy.
Another interesting observation is that, in the preparation of Tat-S-S-dmCocE, only one form of conjugate was generated, and the ratio of unreacted dmCocE was shown to be higher than that in the preparation of LMWP-S-S-dmCocE. One possible explanation for this would be that, according to their amino acid compositions, Tat has a lower isoelectric point (pI = 12.7) than LMWP (pI = 13.5). At pH 7.4, Tat would presumably be less positively-charged compared with LMWP, and therefore would exhibit a weaker electrostatic attraction to the negatively-charged CocE (pI ~4.7). Since the electrostatic attraction between CPP and CocE was known to facilitate the chemical conjugation reaction, a weaker interaction between Tat and CocE might result in less efficient conjugation kinetics. This, in turn, would likely generate fewer types of Tat-S-S-dmCocE variants and leave a higher ratio of the unreacted dmCocE, relative to that in the LMWP-S-S-dmCocE conjugation.
An advantage of the genetic recombination strategy over chemical conjugation is that, genetic recombination provides precise control of the CPP number and location on a CPP-dmCocE fusion protein, as its primary structure is exactly encoded in its DNA sequence. This precise control ensures the reproducibility in production of the CPP-dmCocE fusion proteins. Moreover, according to its three-dimensional structure8, 38, the two termini of dmCocE are exposed outside the surface and also away from the enzyme catalytic center. Hence, the incorporated CPP groups would neither be buried inside, which could lead to a reduced or no cellular uptake, nor impair the enzymatic activity of CocE. This proposition was supported by the elution profile from the heparin affinity column (Figure 6) and the enzyme kinetics (Table 3) of all of the recombinant CPP-dmCocE conjugates. In addition, the E. coli expression system established for dmCocE was also shown to be applicable to generating CPP-dmCocE fusion proteins in soluble forms. In this expression system, the amount of total soluble protein production in cultures expressing dmCocE and CPP-dmCocE fusion proteins were similar. This result suggests that CPP-dmCocE fusion proteins were not toxic or harmful to E. coli, at least no more so than dmCocE itself.
Despite this advantage, two major limitations related to the production of the CPP-dmCocE fusion proteins must be addressed. First, the CPP sequence can only be expressed in tandem; i.e., at the beginning (N-terminus) and/or the end (C-terminus) of the dmCocE sequence. Second, the insertion of CPP sequence might interfere with the expression of dmCocE, lowering its yield or solubility. As seen in Table 2, the expression level of CPP-dmCocE fusion proteins was significantly lower than that of native dmCocE. This reduced expression could be attributed to the codon usage bias in E. coli. Specifically, both Tat and LMWP are composed of a cluster of arginine residues, the amino acid that was known to be encoded in the rarest codons in E. coli and also translated far slower than other amino acids43. Thus, incorporation of Tat or LMWP could hamper the overall expression of CPP-CocE fusion proteins, leading to a lower production yield. The CPP interference was reported to be more pronounced if these arginine-rich sequences were inserted at the N-terminus, where the protein translation starts44. This phenomenon was in the agreement with our observations, in which the expression level of the N-terminal CPP-dmCocE fusion proteins (Tat-N-dmCocE and LMWP-N-dmCocE) was lower than those of the C-terminal ones (dmCocE-C-Tat and dmCocE-C-LMWP). This deficiency may be avoided by re-engineering the arginine codons in the constructs to utilize arginine codons that are optimal for high levels of expression in E. coli.
For both the chemical conjugation and genetic recombination approaches, although attaching Tat or LMWP on dmCocE does not abolish the enzyme activity, this attachment nevertheless reduces the binding affinity, i.e., increases the KM value (see Table 3), of dmCocE for its cocaine substrate. The reduction in substrate binding affinity of CPP-attached dmCocE variants could be attributed to the electrostatic interaction between cocaine and Tat/LMWP at a physiological pH. At pH 7.4, both cocaine and Tat/LMWP are positively charged; thus, the presence of Tat or LMWP could yield a repulsive interaction to cocaine, preventing it from accessing the catalytic center of CocE.
Another noteworthy aspect of the CPP-dmCocE fusion proteins is on their in vitro thermal stability at 37°C. According to the recently published literature, under the in vitro and non-reductive environment, CocE (and dmCocE) undergoes a dimerization process that involves many residues in the N- and C-terminus of CocE35. This dimerization does not impair the enzymatic activity of CocE variants, but seems to play an essential role in their in vitro stabilization. Furthermore, all mutations that stabilize CocE dimerization have shown to significantly improve the thermal stability of CocE6, 7. Given the fact that the CocE dimerization involves largely the N- and C-terminal regions of CocE and has a major influence on the stability, it is reasonable to assume that attaching CPP on either terminus of dmCocE might impair the dimerization, thus reducing the in vitro stability at 37 °C. Interestingly, the N-terminus attached CPP-dmCocE variants displayed a stability profile identical to that of dmCocE within the experimental error (Table 4), whereas the C-terminus attached CPPs were roughly 2-fold less stable than dmCocE. It should be noted that aggregation was reported to be the major mechanism of CocE deactivation35, and it was also a common issue encountered in many CPP-fused, negatively charged proteins45.
Although all of the six FITC-labeled CPP-CocE variants displayed translocations and accumulation inside the HeLa cells, their intracellular distributions differed. As discussed previously, the disulfide bonds in the two chemical CPP-dmCocE conjugates were likely degraded by the elevated reductase activities in the cytosol, leaving the membrane-impermeable dmCocE permanently entrapped, consistent with the even distribution observed inside the cells. In contrast, the four recombinant CPP-dmCocE fusion proteins all displayed a certain degree of nuclear localization, presumably due to the strong DNA binding affinity of CPP. Interestingly, nuclear accumulation was more profound for dmCocE-C-Tat and dmCocE-C-LMWP. It was reported that the polyhistine polymer (n>5) also possessed DNA binding affinity46. Since the CPP groups in these two fusion proteins were attached to the C-termini that were directly adjacent to the 6-histidine tags, the DNA binding affinity of both dmCocE-C-Tat and dmCocE-C-LMWP was enhanced, leading to a more distinct nuclear localization. Furthermore, our results also showed that dmCocE-C-Tat and dmCocE-C-LMWP were not confined inside the endosomes after cell internalization (Figure 8g and 8h). Normally, when CPPs were used to deliver a protein into cells, at least some of CPP-protein conjugates would internalize via endocytosis and therefore localize in the endosomes, forming condensed “bright spots” or vesicular structures in the cytoplasm under microscope27, 34. Hence, for these two CPP-CocE constructs, it would be reasonable to assume that their homogenous cytoplasmic distribution was not related to the types of CPP. To this regard, further investigation of cytoplasmic distribution among different CPP-CocE variants is necessary to fully elucidate the cell translocation mechanism. Regardless of its detailed mechanism, this non-endosomally-confined cytoplasmic delivery is beneficial for the cell-encapsulated CocE, because acidic pH and presence of various proteolytic enzymes inside the endosomes47 would degrade the encapsulated CocE rapidly. Clearly, a homogenous, cytosol-distributed CocE would be more preferable for in vivo cocaine detoxification.
Table 5 summarizes findings of all of the six CPP-dmCocE constructs created in this research. The LMWP-S-S-dmCocE chemical conjugate showed a homogenous cytoplasmic distribution and the highest cocaine-hydrolyzing activity. However, the low production yield (3.9 mg per batch) would hamper its potential for clinical application. On the other hand, the dmCocE-C-Tat fusion protein also displayed a homogenous cytoplasmic distribution. Although its cocaine-hydrolyzing activity ranked as the second highest, it nonetheless gave the largest production yield (73 mg per batch), which was about 20-fold higher than LMWP-S-S-dmCocE. Also noteworthy CPP-CocE variant is Tat-N-dmCocE. It displayed a half-life (5.76 hour) that is 1.3-fold higher over that of DMCocE and almost identical to the most stable CocE variant reported to date7. Based on a balanced consideration of essentials for clinical application, including parameters such as high cocaine-hydrolyzing efficiency, large production yield, major cytoplasmic distribution, etc., the dmCocE-C-Tat fusion protein seems to be the most ideal candidate from this investigation.
Table 5.
In vitro characteristics and cellular uptake behaviors of CPP-dmCocE variants.
| Yield in Mass Quantity Per Batch (mg) |
% Efficiency Compared with dmCocEb |
In Vitro t1/2 at 37 °C (hr) |
Cytoplasmic Distribution |
|
|---|---|---|---|---|
| Chemical Conjugates | ||||
| Tat-S-S-dmCocE | 1.1 (±0.1) | n/a | n/a | Condensed |
| LMWP-S-S-dmCocE | 3.9a | 77–85 | n/a | Homogenous |
| Recombinant Fusion Proteins | ||||
| Tat-N-dmCocE | 32.7 (±0.2) | 53 | 5.76 (±1.45) | Condensed |
| LMWP-N-dmCocE | 7.3 (±0.1) | 45 | 4.83 (±1.78) | Condensed |
| dmCocE-C-Tat | 73.3 (±2.3) | 50 | 1.66 (±0.39) | Homogenous |
| dmCocE-C-LMWP | 10.3 (±0.3) | 49 | 2.98 (±0.57) | Homogenous |
Based on the results presented in this research, further studies that involve the applications of cell-permeable CPP-CocE products in established, clinically relevant mouse models8, 11 are currently underway in our laboratories. One potential clinical application of CPP-CocE is CocE-encapsulated RBC. Owing to the enucleated and relatively inert intracellular environment of RBC, the CocE encapsulated inside the RBC should be even more stable and therefore exhibit the same “life-span” as the host RBC. Normally, human RBC have a life-span of 120 days48, which is significantly longer than the half-life of dmCocE (~4.5 hours) and the life-span of any known synthetic drug carrier. In our previous publication, we already confirmed the feasibility of this RBC encapsulation strategy by using the clinical protein drug asparaginase as an example11. Another potential application of CPP-CocE products is through nasal delivery. The CPP-CocE internalized in the nasal epithelial cells would escape proteases and capillary clearance on the nasal mucous membrane, and therefore can remain active. Furthermore, the CPP-CocE internalized in the nasal cells would gain an opportunity to enter the CNS via the olfactory region49. A proof of concept was provided by Janda’s group in 2004, who displayed a cocaine-binding protein on the surface of a CNS-penetrating bacteriophage50. Through an intranasal administration, this construct has shown the ability to enter the CNS, sequester cocaine in the brain and block its psychoactive effects. Our preliminary observations also indicated that a CPP-conjugated protein can cross the nasal mucosal barrier to reach the brain target (unpublished data).
Overall, this research demonstrates that the attachment of a CPP group enables CocE to be cell permeable while maintaining the enzymatic activity. More importantly, this work also reveals that different ways of CPP attachment can potentially influence a variety of in vitro properties, including the cellular uptake phenotype, of the CPP-attached CocE. Findings from this research are not only useful for developing other new CPP-CocE constructs, but also valuable for establishing a non-disruptive cell-encapsulation technology for other protein therapeutics that are known to be immunogenic or suffer from rapid extracellular degradation for direct clinical application.
Supplementary Material
Acknowledgements
This work was supported by NIH grant R01DA021416, Grant R31-2008-000-10103-01 from the World Class University (WCU) project of South Korea, and the National Basic Research Program of China (973 Program) 2007CB935800. Victor C. Yang is currently a participating faculty member in the Department of Molecular Medicine and Biopharmaceutical Sciences, Seoul National University, South Korea. The authors would also like to thank Dr. Christine Feak at the University of Michigan for her assistance in reviewing the manuscript.
Abbreviations
- CPP
cell penetrating peptides
- CocE
bacterial cocaine esterase
- dmCocE
T172R/G173Q-CocE
- DIC
differential interference contrast
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- E. coli
Escherichia coli
- FPLC
fast protein liquid chromatography
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- HBSS
Hank’s balanced salt solution
- IPTG
isopropyl-β-thiogalactopyranoside
- LMWP
low molecular weight protamine
- MEM-α
minimum essential medium α
- MRT
mean residence time
- PCR
polymerase chain reaction
- PEG
polyethylene glycol
- P2T
Pyridine-2-thione
- RBC
red blood cells
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SPDP
N-succinimidyl 3-(2-pyridyldithio)-propionate
- t1/2
half-life
References
- 1.United Nations Office on Drugs and Crime (UNODC) World Drug Report 2011. 2011:85–126.
- 2.Boghdadi MS, Henning RJ. Cocaine: pathophysiology and clinical toxicology. Heart Lung. 1997;26(6):466–483. doi: 10.1016/s0147-9563(97)90040-6. [DOI] [PubMed] [Google Scholar]
- 3.Bresler MM, Rosser SJ, Basran A, Bruce NC. Gene cloning and nucleotide sequencing and properties of a cocaine esterase from Rhodococcus sp. strain MB1. Appl Environ Microbiol. 2000;66(3):904–908. doi: 10.1128/aem.66.3.904-908.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner JM, Larsen NA, Basran A, Barbas CF, 3rd, Bruce NC, Wilson IA, Lerner RA. Biochemical characterization and structural analysis of a highly proficient cocaine esterase. Biochemistry. 2002;41(41):12297–12307. doi: 10.1021/bi026131p. [DOI] [PubMed] [Google Scholar]
- 5.Cooper ZD, Narasimhan D, Sunahara RK, Mierzejewski P, Jutkiewicz EM, Larsen NA, Wilson IA, Landry DW, Woods JH. Rapid and Robust Protection against Cocaine-Induced Lethality in Rats by the Bacterial Cocaine Esterase. Mol Pharmacol. 2006;70(6):1885–1891. doi: 10.1124/mol.106.025999. [DOI] [PubMed] [Google Scholar]
- 6.Gao D, Narasimhan DL, Macdonald J, Brim R, Ko MC, Landry DW, Woods JH, Sunahara RK, Zhan CG. Thermostable Variants of Cocaine Esterase for Long-Time Protection against Cocaine Toxicity. Mol Pharmacol. 2008;75(2):318–323. doi: 10.1124/mol.108.049486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brim RL, Nance MR, Youngstrom DW, Narasimhan D, Zhan CG, Tesmer JJG, Sunahara RK, Woods JH. A Thermally Stable Form of Bacterial Cocaine Esterase: A Potential Therapeutic Agent for Treatment of Cocaine Abuse. Mol Pharmacol. 2010;77(4):593–600. doi: 10.1124/mol.109.060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park J-B, Kwon YM, Lee T-Y, Brim R, Ko M-C, Sunahara RK, Woods JH, Yang VC. PEGylation of bacterial cocaine esterase for protection against protease digestion and immunogenicity. J Control Release. 2010;142(2):174–179. doi: 10.1016/j.jconrel.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beutler E, Dale GL, Guinto DE, Kuhl W. Enzyme replacement therapy in Gaucher's disease: preliminary clinical trial of a new enzyme preparation. Proc Natl Acad Sci U S A. 1977;74(10):4620–4623. doi: 10.1073/pnas.74.10.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muthuvel A, Rajamani R, Manikandan S, Sheeladevi R. Detoxification of formate by formate dehydrogenase-loaded erythrocytes and carbicarb in folate-deficient methanol-intoxicated rats. Clin Chim Acta. 2006;367(1–2):162–169. doi: 10.1016/j.cca.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Kwon YM, Chung HS, Moon C, Yockman J, Park YJ, Gitlin SD, David AE, Yang VC. L-Asparaginase encapsulated intact erythrocytes for treatment of acute lymphoblastic leukemia (ALL) J Control Release. 2009;139(3):182–189. doi: 10.1016/j.jconrel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutierrez Millan C, Marinero ML, Castaneda AZ, Lanao JM. Drug, enzyme and peptide delivery using erythrocytes as carriers. J Control Release. 2004;95(1):27–49. doi: 10.1016/j.jconrel.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 13.Othman AA, Syed SA, Newman AH, Eddington ND. Transport, metabolism, and in vivo population pharmacokinetics of the chloro benztropine analogs, a class of compounds extensively evaluated in animal models of drug abuse. J Pharmacol Exp Ther. 2007;320(1):344–353. doi: 10.1124/jpet.106.111245. [DOI] [PubMed] [Google Scholar]
- 14.Javaid JI, Dekirmenjian H, Davis JM, Schuster CR. Determination of cocaine in human urine, plasma and red blood cells by gas-liquid chromatography. J Chromatogr. 1978;152(1):105–113. doi: 10.1016/s0021-9673(00)85339-x. [DOI] [PubMed] [Google Scholar]
- 15.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1–3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 16.Collins GT, Brim RL, Narasimhan D, Ko MC, Sunahara RK, Zhan CG, Woods JH. Cocaine esterase prevents cocaine-induced toxicity and the ongoing intravenous self-administration of cocaine in rats. J Pharmacol Exp Ther. 2009;331(2):445–455. doi: 10.1124/jpet.108.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fawell S, Seery J, Daikh Y, Moore C, Chen LL, Pepinsky B, Barsoum J. Tat-mediated delivery of heterologous proteins into cells. Proc Natl Acad Sci U S A. 1994;91(2):664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallbrink M, Floren A, Elmquist A, Pooga M, Bartfai T, Langel U. Cargo delivery kinetics of cell-penetrating peptides. Biochim Biophys Acta. 2001;1515(2):101–109. doi: 10.1016/s0005-2736(01)00398-4. [DOI] [PubMed] [Google Scholar]
- 19.Heitz F, Morris MC, Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Br J Pharmacol. 2009 doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vives E, Brodin P, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J Biol Chem. 1997;272(25):16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Harrison SD. Cell-penetrating peptides in drug development: enabling intracellular targets. Biochem Soc Trans. 2007;35(Pt 4):821–825. doi: 10.1042/BST0350821. [DOI] [PubMed] [Google Scholar]
- 22.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285(5433):1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 23.Leifert JA, Harkins S, Whitton JL. Full-length proteins attached to the HIV tat protein transduction domain are neither transduced between cells, nor exhibit enhanced immunogenicity. Gene Ther. 2002;9(21):1422–1428. doi: 10.1038/sj.gt.3301819. [DOI] [PubMed] [Google Scholar]
- 24.Byun Y, Singh VK, Yang VC. Low molecular weight protamine: a potential nontoxic heparin antagonist. Thromb Res. 1999;94(1):53–61. doi: 10.1016/s0049-3848(98)00201-1. [DOI] [PubMed] [Google Scholar]
- 25.Park YJ, Liang JF, Ko KS, Kim SW, Yang VC. Low molecular weight protamine as an efficient and nontoxic gene carrier: in vitro study. J Gene Med. 2003;5(8):700–711. doi: 10.1002/jgm.402. [DOI] [PubMed] [Google Scholar]
- 26.Park YS, Huang Y, Park YJ, David AE, White L, He H, Chung HS, Yang VC. Specific down regulation of 3T3-L1 adipocyte differentiation by cell-permeable antisense HIF1α-oligonucleotide. J Control Release. 2010;144(1):82–90. doi: 10.1016/j.jconrel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park YJ, Chang LC, Liang JF, Moon C, Chung CP, Yang VC. Nontoxic membrane translocation peptide from protamine, low molecular weight protamine (LMWP), for enhanced intracellular protein delivery: in vitro and in vivo study. Faseb J. 2005;19(11):1555–1557. doi: 10.1096/fj.04-2322fje. [DOI] [PubMed] [Google Scholar]
- 28.Kwon YM, Li YT, Liang JF, Park YJ, Chang LC, Yang VC. PTD-modified ATTEMPTS system for enhanced asparaginase therapy: a proof-of-concept investigation. J Control Release. 2008;130(3):252–258. doi: 10.1016/j.jconrel.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang LC, Lee HF, Yang Z, Yang VC. Low molecular weight protamine (LMWP) as nontoxic heparin/low molecular weight heparin antidote (I): preparation and characterization. AAPS PharmSci. 2001;3(3):E17. doi: 10.1208/ps030317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee LM, Chang LC, Wrobleski S, Wakefield TW, Yang VC. Low molecular weight protamine as nontoxic heparin/low molecular weight heparin antidote (III): preliminary in vivo evaluation of efficacy and toxicity using a canine model. AAPS PharmSci. 2001;3(3):E19. doi: 10.1208/ps030319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang JF, Zhen L, Chang LC, Yang VC. A less toxic heparin antagonist--low molecular weight protamine. Biochemistry (Mosc) 2003;68(1):116–120. doi: 10.1023/a:1022109905487. [DOI] [PubMed] [Google Scholar]
- 32.Tsui B, Singh VK, Liang JF, Yang VC. Reduced reactivity towards anti-protamine antibodies of a low molecular weight protamine analogue. Thromb Res. 2001;101(5):417–420. doi: 10.1016/s0049-3848(00)00427-8. [DOI] [PubMed] [Google Scholar]
- 33.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, Chernomordik LV, Lebleu B. Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J Biol Chem. 2003;278(1):585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 34.Duchardt F, Fotin-Mleczek M, Schwarz H, Fischer R, Brock R. A Comprehensive Model for the Cellular Uptake of Cationic Cell-penetrating Peptides. Traffic. 2007;8(7):848–866. doi: 10.1111/j.1600-0854.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- 35.Narasimhan D, Nance MR, Gao D, Ko MC, Macdonald J, Tamburi P, Yoon D, Landry DM, Woods JH, Zhan CG, Tesmer JJG, Sunahara RK. Structural analysis of thermostabilizing mutations of cocaine esterase. Protein Eng., Des. Sel. 2010;23(7):537–547. doi: 10.1093/protein/gzq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlsson J, Drevin H, Axen R. Protein thiolation and reversible protein-protein conjugation. N-Succinimidyl 3-(2-pyridyldithio)propionate, a new heterobifunctional reagent. Biochem J. 1978;173(3):723–737. doi: 10.1042/bj1730723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawano A, Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 2000;28(16):E78. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larsen NA, Turner JM, Stevens J, Rosser SJ, Basran A, Lerner RA, Bruce NC, Wilson IA. Crystal structure of a bacterial cocaine esterase. Nat Struct Biol. 2002;9(1):17–21. doi: 10.1038/nsb742. [DOI] [PubMed] [Google Scholar]
- 39.Gatley SJ. Activities of the enantiomers of cocaine and some related compounds as substrates and inhibitors of plasma butyrylcholinesterase. Biochem Pharmacol. 1991;41(8):1249–1254. doi: 10.1016/0006-2952(91)90665-r. [DOI] [PubMed] [Google Scholar]
- 40.Rothe R, Liguori L, Villegas-Mendez A, Marques B, Grunwald D, Drouet E, Lenormand JL. Characterization of the Cell-penetrating Properties of the Epstein-Barr Virus ZEBRA trans-Activator. J Biol Chem. 2010;285(26):20224–20233. doi: 10.1074/jbc.M110.101550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziegler A, Seelig J. High affinity of the cell-penetrating peptide HIV-1 Tat-PTD for DNA. Biochemistry. 2007;46(27):8138–8145. doi: 10.1021/bi700416h. [DOI] [PubMed] [Google Scholar]
- 42.Kharidia R, Friedman KA, Liang JF. Improved gene expression using low molecular weight peptides produced from protamine sulfate. Biochemistry (Mosc) 2008;73(10):1162–1168. doi: 10.1134/s0006297908100143. [DOI] [PubMed] [Google Scholar]
- 43.Kane JF. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol. 1995;6(5):494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 44.Goldman E, Rosenberg AH, Zubay G, Studier FW. Consecutive low-usage leucine codons block translation only when near the 5' end of a message in Escherichia coli. J Mol Biol. 1995;245(5):467–473. doi: 10.1006/jmbi.1994.0038. [DOI] [PubMed] [Google Scholar]
- 45.Becker-Hapak M, Dowdy S. Protein transduction: generation of full-length transducible proteins using the TAT system. Current Protocols in Cell Biology. 2003;Vol. 18:20.2.01–20.2.25. doi: 10.1002/0471143030.cb2002s18. [DOI] [PubMed] [Google Scholar]
- 46.Lo SL, Wang S. An endosomolytic Tat peptide produced by incorporation of histidine and cysteine residues as a nonviral vector for DNA transfection. Biomaterials. 2008;29(15):2408–2414. doi: 10.1016/j.biomaterials.2008.01.031. [DOI] [PubMed] [Google Scholar]
- 47.Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med. 2004;10(3):310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 48.Guyton AG, Hall JE. Textbook of Medical Physiology. Philadelphia: W.B. Saunders; 1996. Red blood cells, anemia and polycytemica; pp. 425–433. [Google Scholar]
- 49.Illum L. Is nose-to-brain transport of drugs in man a reality? J Pharm Pharmacol. 2004;56(1):3–17. doi: 10.1211/0022357022539. [DOI] [PubMed] [Google Scholar]
- 50.Carrera MR, Kaufmann GF, Mee JM, Meijler MM, Koob GF, Janda KD. Treating cocaine addiction with viruses. Proc Natl Acad Sci U S A. 2004;101(28):10416–10421. doi: 10.1073/pnas.0403795101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.