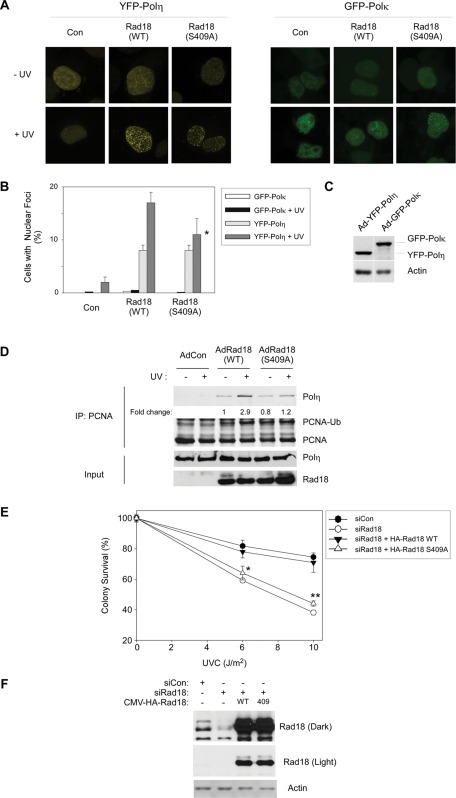
FIGURE 5:
Rad18 S409A does not promote efficient recruitment of Polη to PCNA and fails to complement the UV sensitivity of Rad18-depleted cells. (A) Triplicate cultures of H1299 cells were depleted of endogenous Rad18 and complemented with siRNA-resistant HA-Rad18 (WT) and HA-Rad18 S409A. The Rad18 dependence of UV-induced YFP-Polη and GFP-Polκ redistribution to nuclear foci was determined as described in Materials and Methods. For each experimental condition (performed in triplicate) 60 cells were scored as nuclei containing >20 TLS polymerase foci. Representative images of cells displaying YFP-Polη foci are shown in A. For each treatment, number of cells positive for nuclear foci was expressed as a percentage of YFP/GFP-polymerase–positive cells (B). For GFP-Polκ– and YFP-Polη–expressing cells we performed analysis of variation (ANOVA) followed by Tukey's test to correct for experiment-wise error rates between multiple comparisons. For UV-irradiated cells coexpressing YFP-Polη and Rad18 WT or YFP-Polη and Rad18 S409A, the difference in number of foci was significant (p < 0.05). For GFP-Polκ–expressing cells under these experimental conditions there were no statistically significant differences between groups (p > 0.05). (C) We performed immunoblotting (with anti-GFP antibodies that recognize both GFP and YFP) to confirm that expression levels of GFP-Polκ and YFP-Polη were similar under these experimental conditions. (D) H1299 cells were infected with AdCon, AdRad18 WT, or AdRad18 S409A and treated with UV (20 J/m2) or left untreated for controls. PCNA was immunoprecipitated from the resulting cells, and immunoprecipitates (as well as appropriate input fractions) were analyzed by SDS–PAGE and immunoblotting. Relative levels of PCNA-associated Polη in the adjacent lanes were calculated after densitometric analysis of the Polη immunoblot. (E) H1299 cells were transfected with siRNA directed against the 3′ untranslated region of endogenous Rad18 mRNA or with a nontargeting control siRNA, siCon. The resulting cells were transfected with CMV-driven mammalian expression plasmids encoding HA-Rad18 (WT), HA-Rad18 S409A, or “empty” pAC.CMV vector for control. Control and HA-Rad18–complemented cells were trypsinized and replated in replicate (five replicates per experimental condition) and then treated with varying doses of UV and analyzed for clonogenic survival as described in Materials and Methods. For each siRNA/complementation, the number of surviving colonies from UV-treated cultures was expressed as a percentage of colony number from unirradiated cells. On the survival curves, each data point represents the mean of five replicate determinations, and error bars represent the range. For each dose of UV, we performed ANOVA between groups followed by Tukey's multiple comparison of means test. For cells that received 6 or 10 J/m2 of UV, in ANOVA the p value was <0.0001, which is significant. Results of the Tukey test were as follows: siCon vs. siRad18, p < 0.001 (indicating reduced UV tolerance); siCon vs. siRad18 + WT Rad18, p > 0.05 (indicating no significant difference and therefore full complementation by WT Rad18); siRad18 + Rad18 WT vs. siRad18 + Rad18 S409A, p < 0.001 (indicating significant difference in phenotype between Rad18 WT and Rad18 S409A, and indicated by asterisk and double asterisk for 6 and 10 J/m2, respectively).