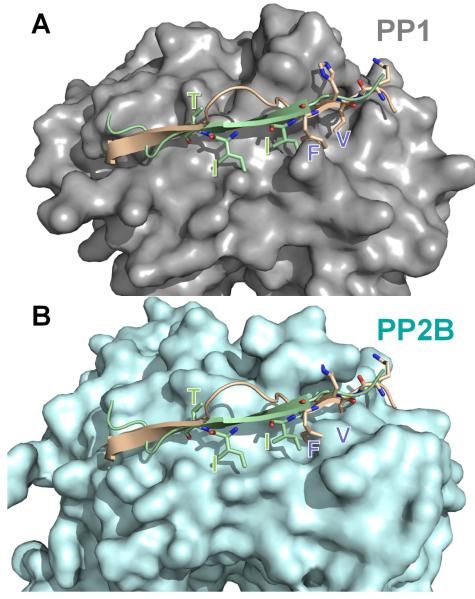
Figure 10. Overlapping, but distinct regulatory and substrate binding sites in PP1/PP2B.
The spinophilin:PP1 (PDB 3EGG) holoenzyme was superimposed onto PP2B:PIVIIT (PDB 2P6B) using PP1 and PP2B. In (A), PP1 (grey) is shown in surface representation with spinophilin (residues 446-461 of spinophilin, which includes the RVxF motif and the β14 binding residues) in beige and the PIVIIT peptide in green. As can be seen, both the PP1 regulator spinophilin, as well as the PP2B substrate mimicking PIVIIT peptide bind to the β14 sheet of PP1/PP2B. (B) Same as in ‘A’ except PP2B (light blue) is now shown in surface representation. The residues that are essential for mediating phosphatase specific interactions are shown in stick representation and labeled in both panels. Comparison of panels ‘A’ and ‘B’ illustrates that the deep hydrophobic pockets that are present in PP1 (RVxF binding pockets) or PP2B (PIVIIT binding pockets) are not present in the other phosphatase. This structural data provides the first molecular insight into the mechanism of specificity regulation between the highly homologous enzymes PP1 and PP2B.