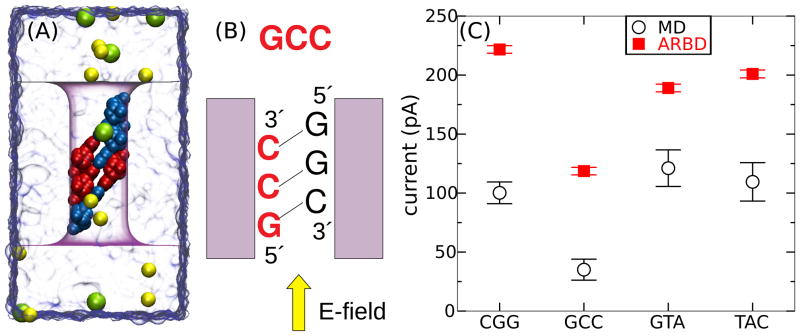
Figure 16.
Ion current through a nanopore blocked by DNA basepair triplets. (A) All-atom MD model of the simulation system. The system is drawn using the same representations as Figure 15A (B) Diagram showing the convention used for naming the sequences. The system shown here is referred to as GCC. (C) Calculated ion current through the pore containing basepair triplets of four different sequences. The MD simulations for CGG and GCC lasted 0.4 μs, while those for GTA and TAC lasted 0.14 μs. The ARBD simulations lasted 16 μs in all four cases. The systematic deviation of the ARBD results is attributable to the higher bulk conductivity of the ARBD solution, Figure 13A, and the high (>2 M) ion concentration in the pore, Figure 11B. The ARBD method qualitatively reproduces large sequence-dependent differences in the currents.