Abstract
Early pregnancy and multiparity are known to reduce the risk of women to develop breast cancer at menopause. The knowledge that the differentiation of the breast induced by the hormones of pregnancy plays a major role in this protection, the present work was performed with the purpose of identifying what differentiation-associated molecular changes persist in the breast until menopause. Core needle biopsies (CNB) obtained from the breast of 42 nulliparous (NP) and 71 parous (P) postmenopausal women were analyzed in morphology, immunocytochemistry and gene expression. Whereas in the NP breast nuclei of epithelial cells were large and euchromatic, in the P breast they were small and hypercromatic, showing strong methylation of istone 3 at lysine 9 and 27. Transcriptomic analysis performed using Affymetrix HG_U133 oligonucleotide arrays revealed that in CNB of the P breast there were 267 upregulated probesets that comprised genes controlling chromatin organization, transcription regulation, splicing machinery, mRNA processing, and noncoding elements including XIST. We concluded that the differentiation process induced by pregnancy is centered in chromatin remodeling and in the mRNA processing reactome, both of which emerge as important regulatory pathways. These are indicative of a safeguard step that maintains the fidelity of the transcription process, becoming the ultimate mechanism mediating the protection of the breast conferred by full term pregnancy.
Keywords: Pregnancy, breast differentiation, H3 methylation, noncoding elements, transcription factors
Introduction
Breast cancer is the most frequently diagnosed cancer in postmenopausal women and the leading cause of cancer death in females worldwide 1. The global incidence of breast cancer has gradually increased over the last few decades 1, 2. Although the reasons of this increase are uncertain, it is known that the breast cancer risk is reduced in women who gave birth to a child before age 24 3, a reduction that is enhanced by breast feeding and multiparity 4, 5. Experimentally it has been demonstrated that the protection conferred by pregnancy is mediated by the differentiation of the breast, a physiological process driven by the complex hormonal milieu created by the placenta and the fetus 6–8. The postulate that the degree of differentiation acquired through an early pregnancy changes the genomic signature that differentiates the lobular structures of early parous women from those of nulliparous women has been demonstrated through the enriched analysis of the genomic profile of breasts of parous and nulliparous postmenopausal and premenopausal women 9, 10 and of rodent models 11, 12. These findings have allowed researchers to demonstrate that significant differences in the expression of genes controlling differentiation and transcription exist between groups that differ in their parity history. These data explain at molecular level the basis of the protective effect of pregnancy and establishes a functional genomic signature of breast cancer risk reduction, confirming a postulate published in 1997 7.
In the present work we present novel findings that emanated from a detailed histological, cytological, immunohistochemical and transcriptomic analysis of breast samples obtained from nulliparous and parous postmenopausal women. Our data demonstrate that the differentiation of the breast induced by an early pregnancy imprints a specific phenotypic and genotypic signature that can be detected in post-menopausal women.
Material and Methods
Data and sample collection
For determining whether the phenotype and the pattern of gene expression differed between nulliparous and parous postmenopausal women, breast tissue was collected from volunteering healthy women residing in Norrbotten County, Sweden, a homogeneously ethnic population of Swedish and Finnish ancestry. Study subjects were recruited between September 2008 and May 2009 at the Mammography Department of Sunderby Hospital, Luleå, Sweden (Study Protocol number 08-020M). The eligibility criteria included women between 50 and 69 years of age, postmenopausal; i.e., lack of menstrual periods for 12 preceding months and elevated circulating levels of follicle stimulating hormone (FSH) (40–250 IU/L), consistent with menopausal condition.
All volunteers signed an informed consent to participate in the study and to donate breast tissues as core needle biopsies (CNB) and blood. Breast core biopsies were taken from the upper outer quadrant of the right or the left breast under radiographic control. From the CNBs obtained from each donor one core was fixed in 70% ethanol for histological and immunohistochemical (IHC) analysis; the two remaining cores were placed in RNAlater® (Ambion) solution for RNA extraction and subsequent transcriptomic analysis. All samples obtained were stripped of any personal identifiers and assigned random numbers that were linked to each subject’s identifiable information which was accessible only to authorized personnel in Sweden 13. All laboratory personnel were blinded to samples’ parity status and other personal information. Unblinding of the parity status after completion of cDNA microarray analysis revealed that volunteer participants belonged to one of the following groups: Parous (P), 71 women that were 23.2 (±4.25) years old at the time of their first pregnancy and 24.0 (±4.6) at the time of delivery of a first full term pregnancy (FTP); this group included women that had been pregnant once or more times (gravida ≥1), and delivered one or more live children (parity ≥1) (G≥1/P≥1); nulligravida nulliparous (NN), 30 women who never became pregnant and therefore never delivered a live child (G0/P0), and gravida-nulliparous (GN), 12 women that became pregnant one or more times, but never delivered a live child (G≥1/P0). Pregnancies in 11 of the women in this group were terminated between the 6th to 12th week of gestation, and one had a miscarriage at the 20th week of pregnancy. Because of the similarities in gene expression levels between NN and GN these two groups were further analyzed as a single group of nulliparous (NP) women.
Methods
Morphological analysis of the architecture of the breast
All ethanol-fixed cores were processed for histopathological and IHC evaluation. All blocks of paraffin-embedded tissues were cut at a thickness of 5 μm, stained with hematoxylin and eosin (HE) and examined under an Olympus BH-2 transmitted light microscope. Each section was evaluated for the presence of mammary ducts and lobules and for determining the ratio between parenchyma and stroma. Specimens lacking epithelial components were not included in the analysis.
Immunohistochemical (IHC) analysis
Fifty cases (21 NP and 29 P) were selected for IHC detection of the following antibodies: cytokeratin (CK) 5/6, clone D5/16 B4, ER-α clone ID5, dilution 1:50; progesterone receptor (PR) (clone PgR 636); smooth muscle antibody (SMA) clone 1A4, and Ki67, all from DakoCytomation; anti-di-methyl-histone H3 (Lys9) (H3K9me2) and tri-methyl histone H3 (Lys27) (H3K27me3) (Cell Signaling Technology Inc.), and cyclin L2 (CCNL2) antibody (Novus Biologicals, Cambridge, UK). All assays were run with appropriate negative and positive controls; incubation and staining procedures were performed following protocols recommended by the manufacturers. Evaluation of IHC reactions was performed by a count of 1,000 cells per case and results were expressed as the percentage of positive nuclei (ER-α, PR, Ki67, H3K9me2, H3K27me3 and CCNL2) or cytoplasm (CK 5/6, SMA) of the total number counted and analyzed by t-test. Cells were evaluated according the intensity of brown staining as strongly positive (+); moderately positive (±), or negative (−).
Image analysis
In normal cells, changes in the chromatin appearance reflect changes in the activation patterns of genes. Features describing the chromatin distribution pattern are referred to as nuclear texture features that are sensitive to the differences between the various descriptive classes of chromatin patterns. In this work we analyzed in hematoxylin-stained slides of breast tissues from four nulliparous and five parous women the nuclear texture features of interphase nuclei of epithelial cells using the Carl Zeiss/Kontron equipment and methodology previously described 14, 15. The threshold low (L) and high (H) levels were defined such that the nuclear images appeared pseudocolorized green and well separated from each other and from the background (in the present case L = ~86 and H = ~200 gray values). Quantitative information on the following geometric, densitometric and textural parameters was obtained: nuclear area (μm2), nuclear perimeter (μm), nuclear feret ratio (= minimum feret/maximal feret), mean gray value per nucleus, standard deviation of the total densitometric values per nucleus or absorbance variability per nucleus (SDtd), entropy and energy 14–17.
Gene analysis
For microarray gene analysis total RNA was isolated from the core biopsy samples using the Qiagen Allprep RNA/DNA Mini Kit (Qiagen, Alameda, CA, USA) according to manufacturer’s instructions. The GeneChip Expression 3′-Amplification Two-Cycle cDNA Synthesis Kit (Affymetrix, Santa Clara, CA) was used for the Affymetrix microarray gene analysis. A total of 169 chips were run; from these, 113 chips, 71 from P and 42 from NP, were selected for the differential expression analysis of parous vs. nulliparous after completion of standard Affymetrix quality control measures (average background, scale factors, percent present calls) and probe-level model (PLM) analysis. Nineteen good quality chips that represented technical replicates were excluded from the P/NP comparison.
Biostatistical analysis
Affymetrix CEL files were pre-processed using RMA 18, which incorporates background adjustment, quantile normalization of probe intensities across arrays, and summarization using the median polish algorithm. To account for between-batch variability in the arrays, the data were adjusted using ComBat, an empirical Bayes framework developed by Johnson et al. 19. A variance filter was applied for removing all probesets with variance across all samples below the first quartile. A total of 18,694 probesets remained for further analysis after filtering. The limma package 20, which uses empirical Bayes methods to moderate standard errors of model coefficients (i.e., log [base 2] fold-changes), was implemented in the R/Bioconductor platform 21 that was used for identifying probesets differentially expressed in the P/NP comparisons. Probesets for downstream analysis from pairwise comparisons were selected using both the p-value of 0.001 from the empirical Bayes moderated t-statistics, and a minimum log2 fold-change of 0.3 threshold as criteria of significance; unless otherwise noted. A heatmap using the RMA expression values and average linkage method to make hierarchical clustering on selected genes was built for depicting the genes that were differentially expressed between parous and nulliparous women. Data mining methods were applied for identifying statistically significant biological processes, pathways and gene networks that were differentially expressed in the P/NP comparisons. Gene ontology (GO) functional categories enriched in differentially expressed genes were identified using conditional hyper-geometric tests in the GOstats package (R/BioConductor). This analysis was carried out independently for up and down regulated genes; a p-value cut off of 0.01 was used to select GO terms.
To identify pathways and associations to other knowledge datasets, Gene Set Enrichment Analysis (GSEA) was performed against the lists of differentially expressed genes. Since we were interested in finding pathways and co-regulated genes, we relaxed the p-value to 0.01 and did not apply any fold change filter to identify pathways from more genes. Pathways obtained from MSigDB (database of gene sets provided by GSEA) were tested for enrichment. Default parameters were chosen, except that the maximum intensity of probes was only selected while collapsing probe sets for a single gene. The gene expression profile obtained was deposited to the Gene Expression Omnibus (GEO) Series GSE26457: (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=zpmfjygscwaemdo&acc=GSE26457).
Gene validation through Real-Time RT-PCR
RNA was reverse-transcribed (RT) using the M-MLV reverse transcriptase (Ambion) and anchored oligo-dT. For each sample, two RT reactions were performed with 50 ng or 12.5 ng of total RNA in 50 μl final. Minus RT controls were included. For one sample arbitrarily chosen as calibrator, a 4-fold 5-points standard curve was performed. Aliquots (5 μl) of cDNA were used for PCR. Real-time Taqman PCR Assays-on-Demand from Applied Biosystems were run using Universal PCR master mix from Applied Biosystems on a 7900 HT instrument. Cycling conditions were 95°C, 15 min followed by 40 (2-step) cycles (95°C, 15 sec; 60°C, 60 sec). The levels of transcripts were expressed as relative quantities to the level in the calibrator. For each sample, the values were averaged and standard deviation of data from two PCR reactions performed with the cDNAs from the two RT reactions. Two-tailed unpaired t-tests were used for analysis of significance; p<0.01 was considered statistically significant.
Results
Architecture of post-menopausal women’s breast
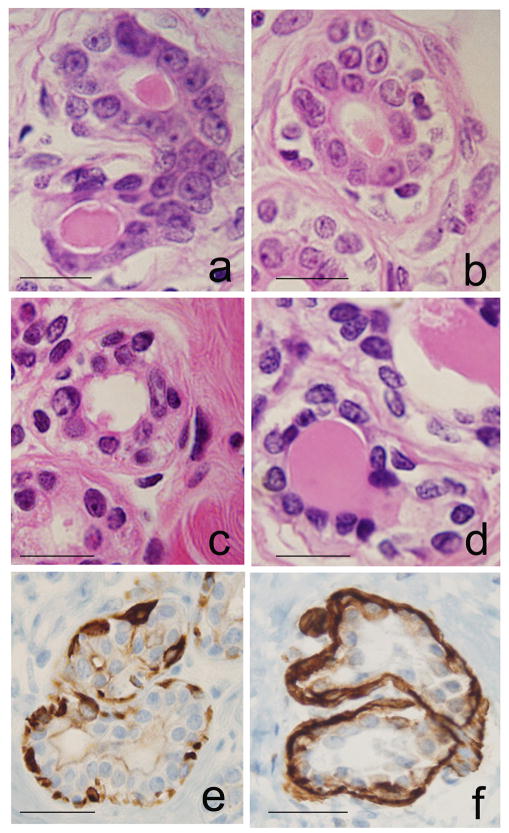
The breast tissues of the parous and nulliparous women contained ducts and lobules type 1 (Lob 1), which were characterized by containing 11.2 ± 6·3 ductules per lobular unit, as previously reported 7–9. Each ductule was composed of an external layer of myoepithelial cells and a monolayer of cuboidal epithelial cells lining a lumen than contained proteinaceous material (Fig. 1a–d). IHC staining revealed that epithelium lining both ducts and ductules consisted of basal cells showing positive reactivity for keratin 5/6 (Fig. 1e) and myoepithelial cells reacting positively with SMA (Fig. 1f); these two markers had similar reactivity in both parous and nulliparous breasts.
Figure 1.
Histological sections of HE-stained ductules in lobules type 1 (Lob 1): a and b, from nulliparous (NP), and c and d from parous (P) women’s breast tissues; e, positive IHC stain for keratin 5/6 in basal cells; f, IHC stain for SMA in myoepithelial cells. DAB with hematoxylin (H) counterstain. Magnification bar: 100 μm.
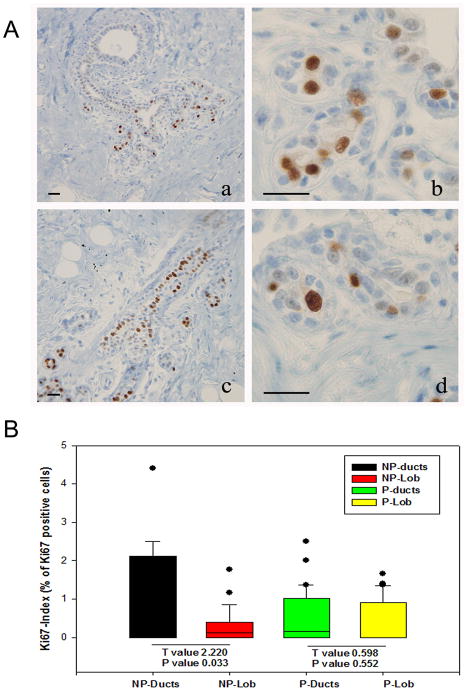
The proliferative activity of the epithelial cells or Ki67 index, was expressed as the percentage of Ki67 positive cells from the total number of cells counted in ducts and Lob 1 of parous (Fig. 2A-a,b) and nulliparous (Fig. 2A-c,d) women’s breast. The Ki67 index in the ductal epithelium of NP breasts’ was significantly higher (P<0.03) than in the Lob 1 of the same tissues and it was also higher than in ducts and Lob 1 of the parous breast (Fig. 2B). In the parous breast, on the other hand, the Ki67 index did not differ between the epithelium of ducts and that of Lob 1 (p>0.55) (Fig. 2B).
Figure 2.
A. Ki 67 immunoreactivity in the nucleus of epithelial cells in ducts (a) and Lob 1 (b) in the parous breast and in ducts (c) and Lob 1 (d) of the nulliparous breast. DAB-H counterstain; magnification bar: 100 μm. B. Box plot of the proliferative activity (Ki67 index) of ductal and Lob 1 epithelial cells. The Ki67 index in ductal epithelial cells of the nulliparous breast (NP-ducts) was significatively higher than in the Lob 1 (NP-Lob) of the same tissues (Paired t-test) (T=2.22; p<0.03), but it did not differ significatively between ducts (P-ducts) and Lob 1 (P-Lob) in the parous breast. The Ki 67 index in NP-ducts was also significative higher than in ducts and Lob 1 of the parous breast.
The ER-α and the PR (data not shown) were equally expressed in epithelial cells lining ducts and Lob 1 of both NP and P breast tissues. The percentage of cells positive for both receptors did not differ between NP and P women’s breasts, as shown in the box plot of ER-α positive cells (Supplemental Figure 1S).
Chromatin pattern in epithelial cells of the nulliparous and parous breast
The study of HE-stained tissues revealed that the population of luminal cells lining ducts and Lob 1 was composed of cells that were characterized by their nuclear appearance into two types: one that contained large and palely stained nuclei with prominent nucleoli (Fig. 1-a,b) and another consisting of small hyper chromatic nuclei (Fig. 1-c,d). The pale staining of the large former nuclei is a feature indicative of a high content of non-condensed euchromatin; these nuclei were called euchromatin-rich nuclei (EUN) (Fig. 1-a,b). The hyperchromasia observed in the latter nuclei was indicative of chromatin condensation and high content of heterochromatin; these nuclei were identified as heterochromatin-rich nucleus (HTN) (Fig. 1-c,d and Fig. 3A). The analysis of the distribution of HTN and EUN cells in histological sections of the breast core biopsies revealed that EUN were more abundant in the NP than in the P breast tissues, whereas the inverse was true for the HTN; these differences were statistically significant (Fig. 3B). We have confirmed the differences between the HTN and EUN depicted in figures 1-a,b vs. 1-c,d, using a quantitative image analysis system. The nuclear size (diameter, area and perimeter) of the EUN as a whole was significantly higher (P<0.05) than that of the HTN in both nulliparous and parous women (Table 1 and Fig. 3B). Differences were also found to be statistically significant (p<0.05) regarding the nuclear shape (nuclear feret ratio) in the breast of nulliparous women, indicating that in these breasts the nuclei of the HTN had a more elongated ellipsoidal shape than the EUN (Table 1). The light absorbance (mean gray values/nucleus) was always greater for EUN than for HTN of both NP and P breasts, either considered as two groups or individually, an indication that under densitometric terms HTN were always more densely stained than EUN. Comparison of the EUN of nulliparous vs. parous breasts revealed significant differences in nuclear size, stainability and densitometric energy, leading us to conclude that epithelial cell nuclei were larger, less stainable and with smaller regions with uniform densitometric intensity in nulliparous breasts. Comparison of the HTN of nulliparous vs. parous breasts revealed significant differences in nuclear diameter (Fig. 3A), perimeter, shape and stainability; cell nuclei showed larger contours and more elongated ellipsoidal shape and they were more stainable in nulliparous breasts (Table 1). These observations indicated that a shift of the EUN cell population to a more densely packed chromatin cell (HTN) had occurred in association with the history of pregnancy as a distinctive pattern of the postmenopausal parous breast.
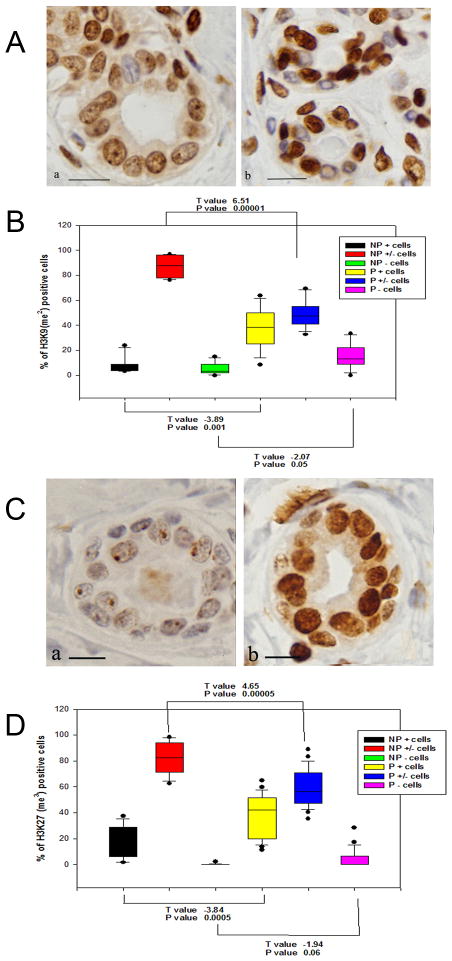
Figure 3.
A. The H3K9(me2) IHC staining is of higher (+) intensity in the nuclei of the P (b) breast than in NP breast (a); a moderately diffuse (±) stain predominates in most of the NP breast. DAB-H counterstain. Magnification bar: 100 μm. B. Box plot shows a significantly higher number of strongly positive cells (+) in P than in NP breasts (p<0.001); moderately positive (±) cells predominate in the NP tissues (p<0.00001), and negative cells are more numerous in the P cells (p<0.05). C. IHC reaction of H3K27(me3) is of higher (+) intensity in the nuclei of the P (b) breast than in NP breast (a), in which the nuclear stain is faint and finely granular, being mostly circumscribed to the nucleoli. DAB-H counterstain. Magnification bar: 100 μm. D. Box plot shows a significantly higher number of strongly positive cells (+) in P than in NP breasts (p<0.0005); weakly to moderately positive (±) cells predominate in the NP tissues (p<0.00005); negative cells are more numerous in the P than in the NP breast cells (p<0.06).
Table 1.
Image analysis geometric, densitometric and textural parameters for hematoxylin-stained interphase nuclei from epithelial cells of postmenopausal nulliparous (NP) and parous (P) women. EUN, euchromatin-rich nucleus; HTN; heterochromatin-rich nucleus; x̄ ± SD, Mean ± standard deviation; ANOVA; NS, no significant..
| Nuclear Characteristics | Reproductive History | |||
|---|---|---|---|---|
| Nulliparous (NP) | Parous (P) | |||
| Chromatin Pattern | EUN x̄ ± SD |
HTN x̄ ± SD |
EUN x̄ ± SD |
HTN x̄ ± SD |
| Area (μm2) | 27.40±7.471 | 16.37±5.082 | 24.38±6.933 | 15.99±4.794 |
| Perimeter (μm) | 24.20±5.341,5 | 18.42±3.722,3 | 22.30±4.793,7 | 16.97±3.014,8 |
| Feret Ratio | 0.72±0.111,9 | 0.62±0.142,10 | 0.72±0.113,11 | 0.71±0.124,12 |
| Mean Gray value/nucleus | 198.64±11.141,13 | 167.96±14.582,14 | 193.85±15.903,15 | 182.90±15.354,16 |
| Entropy | 5.69±0.261,17 | 5.85±0.322,18 | 5.66±0.413,19 | 5.81±0.344,20 |
| Energy | 0.022±0.0051,21 | 0.019±0.0042,22 | 0.024±0.0123,23 | 0.021±0.0064,24 |
1EUN vs. 2HTN (p<0.05); 3EUN vs. 4HTN (p<0.05); EUN 1 vs. 3 (P>0.000); HTN 2 vs. 4 (NS); EUN 5 vs. 7 (P>0.000); HTN 6 vs. 8 (p<0.004); EUN 9 vs. 11 (NS); HTN 10 vs. 12 (p<0.000); EUN 13 vs. 15 (0.000); HTN 14 vs. 16 (p<0.000); EUN 17 vs. 19 (NS); HTN 18 vs. 20 (NS); EUN 21 vs. 23 (p<0.001); HTN 22 vs. 24 (NS).
Since chromatin condensation is part of the process of chromatin remodeling towards gene silencing that is highly regulated by methylation of histones, we verified this phenomenon by IHC incubating NP and P breast tissues with antibodies against histone 3 dimethylated at lysine 9 (H3K9me2) (Figs. 4A and 4B) and trimethylated at lysine 27 (H3K27me3) (Figs. 4C and 4D). The IHC stain revealed that methylation of H3 at both lysine 9 and 27 was increased in the heterochromatin condensed nuclei of epithelial cells of the parous breast (Figs. 4A-b and 4C-b) when compared to the euchromatin rich nuclei of the nulliparous breast (Figs. 4A-a and 4C-a). In the nulliparous breast the reactivity in individual cells was less intense and the number of positive cells was significantly lower (Fig. 4B and 4D). These variations in chromatin reorganization were supported by the upregulation of CBX3, CHD2, L3MBTL, and EZH2 genes controlling this process, as depicted in Table 2.A and Figure 5.
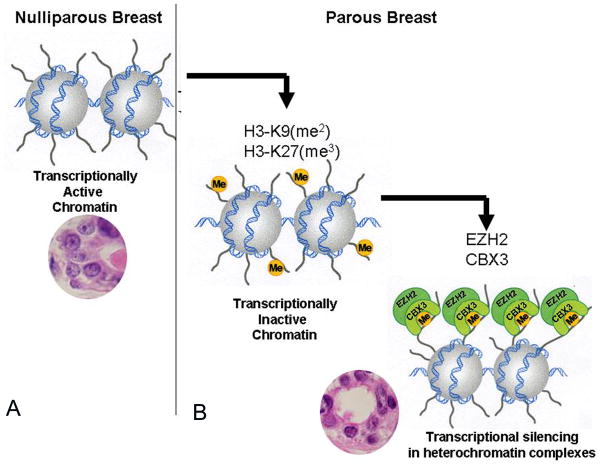
Figure 4.
A. Transcriptionally active chromatin is predominantly expressed in the euchromatin-rich nuclei (EUN) of the nulliparous women’s breast. B. Transcriptionally inactive chromatin is more frequently found in the heterochromatin-rich nuclei (HTN) of the parous breast; its presence is associated with histone 3 methylation at lysines 9 and 27, and transcriptional silencing in heterochromatin complexes.
Table 2.
Genes displaying differential expression between parous and nulliparous breast tissues classified according to biological processes
| Symbol | Log Ratio | P value | Gene Name |
|---|---|---|---|
| Apoptosis (GO:0006915;GO:0006917;GO:0008624;GO:0042981) | |||
|
| |||
| CASP4 | 0.37 | 0.0003 | caspase 4, apoptosis-related cysteine peptidase |
| RUNX3 | 0.36 | 0.0000 | runt-related transcription factor 3 |
| LUC7L3 | 0.34 | 0.0002 | LUC7-like 3 (S. cerevisiae) |
| ELMO3 | 0.30 | 0.0003 | engulfment and cell motility 3 |
|
| |||
| DNA repair (GO:0006281; GO:0006284) | |||
|
| |||
| SFPQ | 0.46 | 0.0002 | splicing factor proline/glutamine-rich |
| MBD4 | 0.36 | 0.0003 | methyl-CpG binding domain protein 4 |
| RBBP8 | 0.32 | 0.0000 | retinoblastoma binding protein 8 |
|
| |||
| Cell adhesion (GO:0007155; GO:0030155) | |||
|
| |||
| NRXN1 | 0.60 | 0.0001 | neurexin 1 |
| DSC3 | 0.51 | 0.0000 | desmocollin 3 |
| COL27A1 | 0.44 | 0.0002 | collagen, type XXVII, alpha 1 |
| PNN | 0.37 | 0.0001 | pinin, desmosome associated protein |
| COL4A6 | 0.36 | 0.0008 | collagen, type IV, alpha 6 |
| LAMC2 | 0.34 | 0.0008 | laminin, gamma 2 |
| COL7A1 | 0.33 | 0.0002 | collagen, type VII, alpha 1 |
| COL16A1 | 0.31 | 0.0000 | collagen, type XVI, alpha 1 |
| LAMA3 | 0.30 | 0.0008 | laminin, alpha 3 |
|
| |||
| Cell cycle (GO:0000075; GO:0007049; GO:0045786) | |||
|
| |||
| SYCP2 | 0.45 | 0.0000 | synaptonemal complex protein 2 |
| PNN | 0.37 | 0.0001 | pinin, desmosome associated protein |
| RUNX3 | 0.36 | 0.0000 | runt-related transcription factor 3 |
| RBBP8 | 0.32 | 0.0000 | retinoblastoma binding protein 8 |
|
| |||
| Cell differentiation (GO:0001709; GO:0030154; GO:0030216) | |||
|
| |||
| MGP | 0.53 | 0.0003 | matrix Gla protein |
| KRT5 | 0.41 | 0.0002 | keratin 5 |
| GATA3 | 0.35 | 0.0009 | GATA binding protein 3 |
| LAMA3 | 0.30 | 0.0008 | laminin, alpha 3 |
|
| |||
| Cell proliferation (GO:0008283; GO:0008284; GO:0008285; GO:0042127; GO:0050679; GO:0050680) | |||
|
| |||
| PTN | 0.67 | 0.0002 | Pleiotrophin |
| KRT5 | 0.41 | 0.0002 | keratin 5 |
| RUNX3 | 0.36 | 0.0000 | runt-related transcription factor 3 |
| IL28RA | 0.34 | 0.0003 | interleukin 28 receptor, alpha (interferon, lambda receptor) |
| CDCA7 | 0.31 | 0.0005 | cell division cycle associated 7 |
|
| |||
| Cell motility (GO:0006928; GO:0030334) | |||
|
| |||
| DNALI1 | 0.37 | 0.0001 | dynein, axonemal, light intermediate chain 1 |
| LAMA3 | 0.30 | 0.0008 | laminin, alpha 3 |
|
| |||
| G-protein coupled receptor pathway (GO:0007186) | |||
|
| |||
| OXTR | 0.54 | 0.0006 | oxytocin receptor |
|
| |||
| RNA metabolic process (GO:0000398; GO:0001510; GO:0006376; GO:0006396; GO:0006397; GO:0006401; GO:0008380) | |||
|
| |||
| METTL3 | 0.69 | 0.0000 | methyltransferase like 3 |
| HNRPDL | 0.65 | 0.0001 | heterogeneous nuclear ribonucleoprotein D-like |
| HNRNPD | 0.59 | 0.0003 | heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| HNRNPA2B1 | 0.56 | 0.0003 | heterogeneous nuclear ribonucleoprotein A2/B1 |
| SFPQ | 0.47 | 0.0006 | splicing factor proline/glutamine-rich |
| RBM25 | 0.38 | 0.0009 | RNA binding motif protein 25 |
| RBMX | 0.38 | 0.0000 | RNA binding motif protein, X-linked |
| LUC7L3 | 0.34 | 0.0002 | LUC7-like 3 (S. cerevisiae) |
| SFRS1 | 0.30 | 0.0001 | splicing factor, arginine/serine-rich 1 |
|
| |||
| RNA transport (GO:0050658) | |||
|
| |||
| HNRNPA2B1 | 0.56 | 0.0003 | heterogeneous nuclear ribonucleoprotein A2/B1 |
|
| |||
| Transcription (GO:0006350; GO:0006355; GO:0006357; GO:0006366; GO:0016481; GO:0045449; GO:0045893; GO:0045941) | |||
|
| |||
| HNRPDL | 0.65 | 0.0001 | heterogeneous nuclear ribonucleoprotein D-like |
| HNRNPD | 0.59 | 0.0003 | heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| CBX3 | 0.53 | 0.0003 | chromobox homolog 3 (HP1 gamma homolog, Drosophila) |
| NFKBIZ | 0.48 | 0.0001 | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| FUBP1 | 0.47 | 0.0002 | far upstream element (FUSE) binding protein 1 |
| SFPQ | 0.47 | 0.0006 | splicing factor proline/glutamine-rich |
| EZH2 | 0.44 | 0.0000 | enhancer of zeste homolog 2 (Drosophila) |
| ZNF207 | 0.41 | 0.0007 | zinc finger protein 207 |
| ZNF711 | 0.41 | 0.0003 | zinc finger protein 711 |
| GATA3 | 0.38 | 0.0009 | GATA binding protein 3 |
| PNN | 0.37 | 0.0003 | pinin, desmosome associated protein |
| ZNF107 | 0.37 | 0.0001 | zinc finger protein 107 |
| RUNX3 | 0.36 | 0.0000 | runt-related transcription factor 3 |
| CCNL1 | 0.35 | 0.0009 | cyclin L1 |
| ZNF692 | 0.34 | 0.0000 | zinc finger protein 692 |
| CHD2 | 0.33 | 0.0001 | chromodomain helicase DNA binding protein 2 |
| RBBP8 | 0.32 | 0.0000 | retinoblastoma binding protein 8 |
| ZNF789 | 0.32 | 0.0005 | zinc finger protein 789 |
| CDCA7 | 0.31 | 0.0005 | cell division cycle associated 7 |
|
| |||
| Chromatin organization (GO:0006333; GO:0006338) | |||
|
| |||
| CBX3 | 0.53 | 0.0003 | chromobox homolog 3 (HP1 gamma homolog, Drosophila) |
| CHD2 | 0.33 | 0.0001 | chromodomain helicase DNA binding protein 2 |
|
| |||
| Cell division (GO:0051301) | |||
|
| |||
| SYCP2 | 0.45 | 0.0000 | synaptonemal complex protein 2 |
|
| |||
| DNA metabolic process (GO:0006139; GO:0006260; GO:0006310; GO:0015074) | |||
|
| |||
| METTL3 | 0.69 | 0.0000 | methyltransferase like 3 |
| SFPQ | 0.46 | 0.0002 | splicing factor proline/glutamine-rich |
| GOLGA2B | 0.32 | 0.0001 | golgin A2 family, member B |
|
| |||
| Lactation (GO:0007595) | |||
|
| |||
| OXTR | 0.54 | 0.0006 | oxytocin receptor |
|
| |||
| Apoptosis (GO:0006917) | |||
|
| |||
| SOS1 | −0.23 | 0.0040 | son of sevenless homolog 1 |
|
| |||
| Cell adhesion (GO:0007155; GO:0030155) | |||
|
| |||
| PDZD2 | −0.35 | 0.0004 | PDZ domain containing 2 |
|
| |||
| Cell proliferation (GO:0008283; GO:0008284; GO:0008285; GO:0042127; GO:0050679; GO:0050680) | |||
|
| |||
| IGF1 | −0.35 | 0.0002 | insulin-like growth factor 1 (somatomedin C) |
|
| |||
| Cell motility (GO:0006928; GO:0030334) | |||
|
| |||
| IGF1 | −0.35 | 0.0002 | insulin-like growth factor 1 (somatomedin C) |
|
| |||
| G-protein coupled receptor pathway (GO:0007186) | |||
|
| |||
| RASD1 | −0.31 | 0.0009 | RAS, dexamethasone-induced 1 |
|
| |||
| Transcription (GO:0006350; GO:0006355; GO:0006357; GO:0006366; GO:0016481; GO:0045449; GO:0045893; GO:0045941) | |||
|
| |||
| SOX17 | −0.28 | 0.0026 | SRY (sex determining region Y)-box 17 |
| EBF1 | −0.33 | 0.0005 | early B-cell factor 1 |
|
| |||
| DNA metabolic process (GO:0006139; GO:0006260; GO:0006310; GO:0015074) | |||
|
| |||
| IGF1 | −0.35 | 0.0002 | insulin-like growth factor 1 (somatomedin C) |
|
| |||
| Insulin-like growth factor receptor signaling pathway (GO:0043568) | |||
| IGF1 | −0.35 | 0.0002 | insulin-like growth factor 1 (somatomedin C) |
Transcriptomic differences
Analysis of P and NP microarrays revealed that there were 305 differentially expressed probesets, 267 up- and 38 down-regulated, corresponding to 208 distinct genes between these two groups. From these 267 genes we selected those that described biological processes that were representative of the transcriptomic differences between the parous and the nulliparous breasts. Using bioinformatics based analysis of microarray data we found that the biological processes involving the splicing machinery and mRNA processing were prevalent in the parous breast and were represented by the following upregulated genes: LUC7L3, SFRS1, HNRNPA2B1, HNRNPD, RBM25, SFRS5, METTL3, HNRNPDL, and SFPQ (Table 2A). Transcription regulation and chromatin organization were also highly represented in the parous breast by the upregulation of CBX3, EBF1, GATA3, RBBP8, CCNL1, CCNL2, CDCA7, EZH2, FUBP1, NFKBIZ, RUNX3, ZNF107, ZNF207, ZNF692, ZNF711, ZNF789, CDCA7, and ZNF692 (Table 2A). The parous breast also expressed upregulation of six non coding regions that included XIST, MALAT-1 (or NEAT2) and NEAT1 (Table 2C).
Genes that were downregulated in the parous breast represented transcription regulation, encompassing CBL, FHL5, NFATC3, NCR3C1, TCF7L2, and a set of genes that were involved in IGF-like growth factor signaling, somatic stem cell maintenance, muscle cell differentiation and apoptosis, such as IGF1, RASD1, EBF1, SOX 1, SOX6, SOX 17, RALGAPA2 and ABHD5 (Table 2B). The level of expression was confirmed to be differentially expressed between nulliparous and parous breast tissues by real time RT-PCR for the following genes: CREBZF, XIST, MALAT1, NEAT1, CCNL2, GATA3, DDX17, HNRPDL, SOX6, SNHG12, SOX 17 and C1orf168. In addition to the level of expression, the localization of the alternative splicing regulator cyclin-cyclin L2 protein (CCNL2) 22 was verified by IHC. CCNL2 protein was expressed in the nucleus of epithelial cells in breast tissues from NP and P women, although the level of expression was significantly higher in Lob 1 in the parous breast when compared with similar structures found in the breast of nulliparous women (Results not shown). These observations confirmed the localization of this gene product in the splicing factor compartment (nuclear speckles) 23.
Discussion
Our previous studies have in great part clarified the role of pregnancy-induced breast differentiation in the reduction in breast cancer risk, as well as the identification of Lob 1 or the terminal ductal lobular unit (TDLU) as the site of origin of breast cancer 6–9, 24. The morphological, physiological and genomic changes resulting from pregnancy and hormonally-induced differentiation of the breast and their influence on breast cancer risk have been addressed in previous publications 6–9, 24, 25. Our observations that during the post-menopausal years the breast of both parous and nulliparous women contains preponderantly Lob 1 and the fact that nulliparous women are at higher risk of developing breast cancer than parous women indicate that Lob 1 in these two groups of women either differ biologically, or exhibit different susceptibility to carcinogenesis 25. For clarifying this concept the present manuscript places major emphasis on the changes in cell types and increases in chromatin condensation as novel markers for defining the concept of differentiation in the adult breast. These findings confirm the universality of the histone 3 methylation in lysines 9 and 27 during differentiation, since a similar phenomenon has been described to occur during embryonic stem cell (ESC) differentiation 26. The observed chromatin changes in parous epithelial cells are complemented by the expression of genes related to increasing cell adhesion and differentiation, such as NRXN1, DSC3, COL27A1, PNN, COL4A6, LAMC2, COL7A1, COL16A1, and LAMA3, and MGP, KRT5, GATA3 and LAMA3, respectively.,
In contrast to the findings of Asztalos et al.10 of downregulation of the expression of ER-α following recent (<2 years) and distant (5 to 10 years) pregnancies in premenopausal women, our current genomic and IHC study did not reveal differences in the level of expression of ER-α in the epithelial cells of ducts and Lob 1 between parous and nulliparous postmenopausal women. Nevertheless, numerous genes that are regulated downstream by the ER-α were found to be upregulated in the parous breast, supporting a-parity mediated protective effect evident in younger parous women 10 but lasting until menopause. Among the ER-α downstream regulated genes was GATA3, which encodes a protein that belongs to the GATA family of transcription factors that regulates T lymphocyte differentiation and maturation. GATA3 is crucial to mammary gland morphogenesis and differentiation of progenitor cells and a putative tumor suppressor 27. Induction of GATA 3 expression in GATA3-negative undifferentiated carcinoma cells is sufficient to induce tumor differentiation and inhibition of tumor dissemination 28. Therefore, the observation that genes involved in the estrogen receptor regulated pathways are upregulated in the parous breast in spite of the lack of transcriptomic differences in this receptor’s levels between parous and nulliparous postmenopausal breast tissues suggests that they could be under permanent transcriptional modification as a manifestation of a higher degree of cell differentiation.
Studies of breast development under the influence of parity in women and in animal models are in agreement on the pregnancy-induced differentiation of the breast, a process that ultimately becomes manifested as a specific genomic signature in the mammary gland 6–12. Although variations in gene expression among different studies and species are expected, an increase in immune activity, including overexpression of lipopolysaccharide binding protein (LBP/Lbp) has been reported in the post-pregnancy breast of premenopausal women 10 and in the mammary gland of four different strains of rats 12. Interestingly, this response observed in both recently pregnant in distant pregnant groups was not observed in the postmenopausal group reported in this study. These discrepancies might indicate that the upregulation of inflammation/immune response–related genes persists during post-partum involution, but wanes after menopause sets in. However, it cannot be ruled out the possibility that the cyclic hormonal changes occurring during the menstrual/estrus cycle influence the genomic profile of the breast activating pathways that greatly differ from those expressed in the postmenopausal parous women.
Importantly, we found a shift in the cell population of the postmenopausal breast as a manifestation of the reprogramming of the organ after pregnancy. These observations are in agreement with what is observed in the rat mammary gland, which also contains two types of luminal epithelial cells, designated dark (DC) and intermediate (IC) cells, in addition to the myoepithelial cells.29 The DC and IC are equivalent to the HTN and EUN cells described in the present work. DCs increase after pregnancy and lactational involution; whereas the ICs significantly outnumber the DC in ductal hyperplasias and ductal carcinomas 29, 30. Our analysis of nuclear ultrastructural and morphometric parameters of rodent IC have allowed us to differentiate the mammary progenitor stem cell from the cancer stem cells 25, 29, 30. Nuclear morphometric analysis of breast and ovarian carcinomas has confirmed the predictive value of nuclear grade on the progression of premalignant lesions to invasiveness 31–33. Our present findings of a significant decrease in the number of EUN with a subsequent increase in the number of HTN cells expressing specific biomarkers identified at the chromatin and transcriptional levels support the value of morphometric analysis as an adjuvant to molecular studies[Figure 4. A–D]. Our data clearly indicate that there are morphological indications of chromatin remodeling in the parous breast, such as the increase in the number of epithelial cells with condensed chromatin and increased reactivity with anti-H3K9me2 and H3K27me3 antibodies. Histone methylation is a major determinant for the formation of active and inactive regions of the genome and is crucial for the proper programming of the genome during development 34. In the parous breast there is upregulation of transcription factors and chromatin remodeling genes such as CHD2 or chromodomain helicase DNA binding protein 2 and the CBX3 or Chromobox homolog 3, whose products are required for controlling recruitment of protein/protein or DNA/protein interactions. CBX3 is involved in transcriptional silencing in heterochromatin-like complexes, and recognizes and binds H3 tails methylated at lysine 9, leading to epigenetic repression. Two other important genes related to the polycomb group (PcG) protein that are upregulated in the parous breast are the L3MBTL gene or l(3)mbt-like and the histone-lysine N-methyltransferase or EZH2. Members of the PcG form multimeric protein complexes that maintain the transcriptional repressive state of genes over successive cell generations. EZH2 is an enzyme that acts mainly as a gene silencer, performing this role by the addition of three methyl groups to lysine 27 of histone 3, a modification that leads to chromatin condensation 26, 35, 36.
Recent studies indicate that RNA molecules recruit PcG complexes to the locus of transcription or to sites located elsewhere in the genome. An important role has been attributed to noncoding RNAs (ncRNAs) 37. It is possible to postulate that the increased chromatin condensation in the parous breast reported herewith could have been initiated by ncRNAs, a postulate supported by the observed upregulation of several ncRNAs that included nuclear paraspeckle assembly transcript 1 (NEAT1), MALAT-1 (NEAT2), and X inactive specific transcript (XIST) 38, all critical components of the speckles. The expression of MALAT-1 is upregulated by the neurotransmitter oxytocin during lactation, which acts through its specific receptor OTR. It is of interest the fact that both OTR and MALAT-1 remain upregulated in the breast of postmenopausal parous women even in the absence of circulating oxytocin. These observations indicate that in parous women the breast remains actively involved in the RNA metabolism that is necessary for maintaining a state of differentiation. Up-regulation of XIST occurs upon differentiation, resulting in X chromosome inactivation. A ncRNA transcribed from a portion of the Xist gene locus forms hairpin structures that recruit the PRC2 complex to the X-inactivation center X(ic) 39. Transcription of full-length Xist RNA, which forms the same hairpin structures, leads to further PRC2 recruitment and the spread of PcG-mediated repression across the inactive X chromosome. Xist repression, which is often seen in malignancies, also occurs in early embryogenesis and during the acquisition of pluripotency in undifferentiated ES cells by the binding of Nanog, Oct4 and Sox2 directly to the chromatin of the Xist gene 39. It is possible that in the postmenopausal nulliparous breast the upregulation of DDX, Sox 1, Sox 6 and Sox 17, which might be equivalent to Nanog, Oct4 and Sox2, play a direct pivotal role in the repression of XIST transcription. Although this postulate needs to be functionally verified, it is possible that these genes may play a role in controlling XIST in the parous breast. The upregulation of XIST has important implications in the understanding of the differentiation pattern of the parous breast. In recent studies it has been shown that the reprogramming of X-chromosome inactivation during the acquisition of pluripotency in vivo and in vitro is accompanied by the repression of XIST 39–41. Reprogramming experiments have further reinforced the concept that X-inactivation is intimately linked to differentiation and support our findings that XIST is expressed in adult well differentiated cells, participating in the maintenance of gene repression 39–41. There is a relationship between the chromatin remodeling process and post transcriptional control maintained by the spliceosome machinery that is stored in nuclear speckles. Among the components of the spliceosome machinery that are up-regulated in the parous breast are the heterogeneous nuclear ribonucleoproteins HNRPA3, HNRPA2B1, HNRPD and the HNRPU shown in Table 2A. The functional role of these HNRPs in the postmenopausal breast could be implicated in the regulation of mRNA stability, other functions like mammary gland involution 42, acting as negative regulators of telomere length maintenance 43 or regulating the trafficking of mRNA molecules 44. Other members of the spliceosome complex are the small nuclear ribonucleoproteins (snRNPs), which function as suppressors of tumor cell growth and may have major implications as cancer therapeutic targets. Among these we have found that the transcripts regulated by the genes SF3B1, SFRS2, SFRS7, SFRS8, SFRS14, SFRS16, SNRP70,, SNRPB, SNRPA1, PRF3 and PHF5A are overexpressed in the parous breast. Other members of the splicing factor compartment that are localized in the nuclear speckles are CCNL1 and CCNL2. We have demonstrated through immunohistochemistry that CCNL2 protein is overexpressed in the nucleus of epithelial cells composing the Lob 1 of the parous breast. CCNL1 and CCNL2 are transcriptional regulators that participate in the pre-mRNA splicing process and the expression of critical factors leading to cell apoptosis, possibly through the Wnt signal transduction pathway 45, 46, which we found to be down-regulated in the parous breast.
Another component of the spliceosome complex that regulates genes involved in the apoptotic process is the RNA binding motif protein 5 (RBM5). The overexpression of RBM5 retards ascites associated tumor growth and enhances p53-mediated inhibition of cell growth and colony formation 47, 48, mechanisms that could also be operational in the parous breast. The spliceosome plays a critical role in differentiating mouse ESC, and self-renewal, pluripotency and tissue lineage specification of human ESC 49. Post-transcriptional modifications of RNA, including packaging into the nuclear speckles of the breast epithelial cells and recognition by RNA-binding proteins and/or microRNAs are crucial processes in differentiating breast epithelial cells. Although it is known that these regulatory mechanisms decrease the susceptibility of the cell to carcinogenesis, more studies need to be conducted for identifying the specific pathways involved in this process. Data presented here contribute to emphasize the importance of post-transcriptional regulatory mechanisms as a critical component underlying the differentiation of the breast.
In summary, in the present work we clearly demonstrate that the breast of parous postmenopausal women exhibits a specific signature that has been induced by a full term pregnancy. This signature reveals for the first time that the differentiation process is centered in chromatin remodeling and the mRNA processing reactome, which emerge as important regulatory pathways induced by pregnancy. This study is strengthened by the homogeneity of a population restricted to participants of Swedish or Finnish ethnicity residents of the northernmost part of Sweden because it avoids variations in gene expression resulting from differences in ethnicity rather than parity. Nevertheless, it also represents a limitation and emphasizes the need of confirming these results in ethnically different and varied populations. The biological importance of the pathways identified in this specific population cannot be sufficiently emphasized due to the fact that the upregulation of the non-coding sequences that control gene repression and of the genes that control the spliceosomes could represent a safeguard mechanism at genomic and at post-transcriptional level that maintains the fidelity of the transcription process, a phenomenon that could be the ultimate step mediating the protection of the breast conferred by full term pregnancy.
Supplementary Material
The estrogen receptor α (ER-α) immunoreactivity in the nucleus of epithelial cells in ducts and Lob 1 is similar between parous and nulliparous breasts, as shown in the box plot.
Table 3.
Non-protein coding (npc) regions overexpressed in the breast of parous postmenopausal women
| GeneSymbol | Probe ID | Log Ratio | Gene Names |
|---|---|---|---|
| CXorf50B | 242292_at | 0.35 | non-protein coding RNA 246B |
|
| |||
| MALAT1 | 224558_s_at | 0.56 | metastasis associated lung adenocarcinoma transcript 1 |
|
| |||
| MALAT1 | 224558_s_at | 0.56 | metastasis associated lung adenocarcinoma transcript 1 |
|
| |||
| NCRNA00173 | 237591_at | 0.39 | non-protein coding RNA 173 |
|
| |||
| NCRNA00201 | 225786_at | 0.47 | non-protein coding RNA 201 |
| NEAT1 | 224565_at | 0.38 | nuclear paraspeckle assembly transcript 1 |
| NEAT1 | 224566_at | 0.50 | nuclear paraspeckle assembly transcript 1 |
|
| |||
| XIST | 224589_at | 0.39 | X (inactive)-specific transcript |
|
| |||
| XIST | 221728_x_at | 0.57 | X (inactive)-specific transcript |
|
| |||
| XIST | 214218_s_at | 0.57 | X (inactive)-specific transcript |
Novelty.
This article demonstrates that an early pregnancy induces lifetime changes in the breast epithelium which are detected in postmenopausal women as a genomic signature that is characterized by chromatin remodeling and post-transcriptional modifications of RNA. These findings explain at molecular level the mechanisms mediating the protective effect conferred by an early pregnancy.
Impact.
The identification of a specific genomic signature of pregnancy has uncovered a novel tool that will serve as a surrogate biomarker for testing new chemopreventive agents and will significantly advance the field of cancer prevention.
Acknowledgments
The authors thank the women of Norrbotten County, Sweden, for their willing contribution to the project, the staff of the Mammography Department, Sunderby Hospital, Luleå, Sweden, and the excellent technical support provided by the Genomics Facility and the Biostatistics and Bioinformatics Facility at the Fox Chase Cancer Center.
Grant Support
Grant sponsor: Avon Foundation (Women Breast Cancer Research Program); Grant number: 02-2008-034; National Institutes of Health; Grant number: CA006927; São Paulo State Research Foundation (Carl Zeiss/Kontron equipment); Grant number: FAPESP, no. 06/0066-8.
Abbreviations
- CNB
Core needle biopsy
- IHC
Immunohistochemistry
- μm
micrometer
- NN
nulligravida nulliparous
- NP
Nulliparous
- P
Parous
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global Cancer Statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Harford JB. Breast-cancer early detection in low-income and middle-income countries: do what you can versus one size fits all. Lancet Oncol. 2011;12:306–12. doi: 10.1016/S1470-2045(10)70273-4. [DOI] [PubMed] [Google Scholar]
- 3.MacMahon B, Cole P, Lin TM, Lowe CR, Mirra AP, Ravnihar B, Salber EJ, Valaoras VG, Yuasa S. Age at first birth and breast cancer risk. Bull World Health Organ. 1970;43:209–21. [PMC free article] [PubMed] [Google Scholar]
- 4.Hinkula M, Pukkala E, Kyyrönen P, Kauppila A. Grand multiparity and the risk of breast cancer: population-based study in Finland. Cancer Causes Control. 2001;12:491–500. doi: 10.1023/a:1011253527605. [DOI] [PubMed] [Google Scholar]
- 5.Yang XR, Chang-Claude J, Goode L, Couch FJ, Nevanlinna H, Milne RL, Gaudet M, Schmidt MK, Broeks A, Cox A, Fasching PA, Hein R, et al. Associations of breast cancer risk factors with tumor subtypes: a pooled analysis from the Breast Cancer Association Consortium studies. J Natl Cancer Inst. 2011;103:250–63. doi: 10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russo IH, Koszalka M, Russo J. Comparative study of the influence of pregnancy and hormonal treatment on mammary carcinogenesis. Br J Cancer. 1991;64:481–4. doi: 10.1038/bjc.1991.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russo J, Russo IH. Role of differentiation in the pathogenesis and prevention of breast cancer. J Endocrinology. 1997;4:7–21. [Google Scholar]
- 8.Russo J, Russo IH, editors. Molecular basis of breast cancer: Prevention and treatment. Berlin, Heidelberg, New York: Springer-Verlag; 2004. p. 447. [Google Scholar]
- 9.Russo J, Balogh GA, Russo IH. Full-term Pregnancy Induces a Specific Genomic Signature in the Human Breast. Cancer Epidemiol Biomarkers Prev. 2008;17:51–66. doi: 10.1158/1055-9965.EPI-07-0678. [DOI] [PubMed] [Google Scholar]
- 10.Asztalos S, Gann PH, Hayes MK, Nonn L, Beam CA, Dai Y, Wiley EL, Tonetti DA. Gene expression patterns in the human breast after pregnancy. Cancer Prev Res. 2010;3:301–11. doi: 10.1158/1940-6207.CAPR-09-0069. [DOI] [PubMed] [Google Scholar]
- 11.Medina D. Breast cancer: the protective effect of pregnancy. Clin Cancer Res. 2004;10:380S–4S. doi: 10.1158/1078-0432.ccr-031211. [DOI] [PubMed] [Google Scholar]
- 12.D’Cruz CM, Moody SE, Master SR, Hartman JL, Keiper EA, Imielinski MB, Cox JD, Wang JY, Ha SI, Keister BA, Chodosh L. Persistent parity-induced changes in growth factors, TGF-beta3, and differentiation in the rodent mammary gland. Mol Endocrinol. 2002;16:2034–51. doi: 10.1210/me.2002-0073. [DOI] [PubMed] [Google Scholar]
- 13.Belitskaya-Levy I, Zeleniuch-Jacquotte A, Russo J, Russo IH, Bordas P, Ahman J, Afanasyeva Y, Johansson R, Lenner P, Li X, Lopez de Cicco R, Peri S, Ross E, Russo PA, Santucci-Pereira J, Sheriff FS, Slifker M, Hallmans G, Toniolo P, Arslan AA. Characterization of a Genomic Signature of Pregnancy in the Breast. Cancer Prev Res. 2011;4:1–24. doi: 10.1158/1940-6207.CAPR-11-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mello ML, Vidal BC, Russo IH, Lareef MH, Russo J. DNA content and chromatin texture of human breast epithelial cells transformed with 17-beta-estradiol and the estrogen antagonist ICI 182,780 as assessed by image analysis. Mutat Res. 2007;617:1–7. doi: 10.1016/j.mrfmmm.2006.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mello ML, Russo P, Russo J, Vidal BC. Entropy of Feulgen-stained 17-beta-estradiol-transformed human breast epithelial cells as assessed by restriction enzymes and image analysis. Oncol Rep. 2009;21:1483–7. doi: 10.3892/or_00000378. [DOI] [PubMed] [Google Scholar]
- 16.Doudkine A, MaCaulay C, Poulin N, Palcic B. Nuclear texture measurements in image cytometry. Pathologica. 1995;87:286–99. [PubMed] [Google Scholar]
- 17.Isharwal S, Miller MC, Marlow C, Makarov DV, Partin AW, Veltri RW. p300 (histone acetyltransferase) biomarker predicts prostate cancer biochemical recurrence and correlates with changes in epithelia nuclear size and shape. Prostate. 2008;68:1097–104. doi: 10.1002/pros.20772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 19.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 20.Smyth GK. limma: Linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 21.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 23.Herrmann A, Fleischer K, Czajkowska H, Müller-Newen G, Becker W. Characterization of cyclin L1 as an immobile component of the splicing factor compartment. FASEB J. 2007;21:3142–52. doi: 10.1096/fj.07-8377com. [DOI] [PubMed] [Google Scholar]
- 24.Russo J, Rivera R, Russo IH. Influence of age and parity on the development of the human breast. Breast Cancer Res Treat. 1992;23:211–8. doi: 10.1007/BF01833517. [DOI] [PubMed] [Google Scholar]
- 25.Russo IH, Russo J. Pregnancy-induced changes in breast cancer risk. J Mammary Gland Biol Neoplasia. 2011;16:221–33. doi: 10.1007/s10911-011-9228-y. [DOI] [PubMed] [Google Scholar]
- 26.Golob JL, Paige SL, Muskheli V, Pabon L, Murry CE. Chromatin remodeling during mouse and human embryonic stem cell differentiation. Dev Dyn. 2008;237:1389–98. doi: 10.1002/dvdy.21545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson BJ, Giguère V. Meta-analysis of human cancer microarrays reveals GATA3 is integral to the estrogen receptor alpha pathway. Mol Cancer. 2008;4(7):49. doi: 10.1186/1476-4598-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chou J, Provot S, Werb Z. GATA3 in development and cancer differentiation: cells GATA have it! J Cell Physiol. 2010;222:42–9. doi: 10.1002/jcp.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russo IH, Russo J. Mammary Gland Neoplasia in Long-Term Rodent Studies. Environ Health Perspect. 1996;104:938–67. doi: 10.1289/ehp.96104938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russo J, Tait L, Russo IH. Susceptibility of the mammary gland to carcinogenesis: III. The cell of origin of rat mammary carcinoma. Am J Path. 1983;113:50–66. [PMC free article] [PubMed] [Google Scholar]
- 31.Bussolati G, Marchiò C, Gaetano L, Lupo R, Sapino A. Pleomorphism of the nuclear envelope in breast cancer: a new approach to an old problem. J Cell Mol Med. 2008;12:209–18. doi: 10.1111/j.1582-4934.2007.00176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan PH, Goh BB, Chiang G, Bay BH. Correlation of nuclear morphometry with pathologic parameters in ductal carcinoma in situ of the breast. Mod Pathol. 2001;14 :937–41. doi: 10.1038/modpathol.3880415. [DOI] [PubMed] [Google Scholar]
- 33.Palmer JE, Sant Cassia LJ, Irwin CJ, Morris AG, Rollason TP. The prognostic value of nuclear morphometric analysis in serous ovarian carcinoma. Int J Gynecol Cancer. 2008;18:692–701. doi: 10.1111/j.1525-1438.2007.01093.x. [DOI] [PubMed] [Google Scholar]
- 34.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 35.Kubicek S, Schotta G, Lachner M, Sengupta R, Kohlmaier A, Perez-Burgos L, Linderson Y, Martens JH, O’Sullivan RJ, Fodor BD, Yonezawa M, Peters AH, et al. The role of histone modifications in epigenetic transitions during normal and perturbed development. Ernst Schering Res Found Workshop. 2006;57:1–27. doi: 10.1007/3-540-37633-x_1. [DOI] [PubMed] [Google Scholar]
- 36.Lin W, Dent SY. Functions of histone-modifying enzymes in development. Curr Opin Genet Dev. 2006;16:137–42. doi: 10.1016/j.gde.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 37.Matthew GG, Young RA. Repressive transcription. Science. 2010;329:150–1. doi: 10.1126/science.1193995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erwin JA, Lee JT. Characterization of X-chromosome inactivation status in human pluripotent stem cells. Curr Protoc Stem Cell Biol. 2010;Chapter 1(Unit 1B.6) doi: 10.1002/9780470151808.sc01b06s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Navarro P, Chambers I, Karwacki-Neisius V, Chureau C, Morey C, Rougeulle C, Avner P. Molecular coupling of Xist regulation and pluripotency. Science. 2008;321:1693–5. doi: 10.1126/science.1160952. [DOI] [PubMed] [Google Scholar]
- 40.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–62. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 41.Lee JT. Characterization of X-chromosome inactivation status in human pluripotent stem cells. Curr Protoc Stem Cell Biol. 2010;Chapter 1(Unit 1B.6) doi: 10.1002/9780470151808.sc01b06s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taga Y, Miyoshi M, Okajima T, Matsuda T, Nadano D. Identification of heterogeneous nuclear ribonucleoprotein A/B as a cytoplasmic mRNA-binding protein in early involution of the mouse mammary gland. Cell Biochem Funct. 2010;28:321–8. doi: 10.1002/cbf.1662. [DOI] [PubMed] [Google Scholar]
- 43.Huang PR, Hung SC, Wang TC. Telomeric DNA-binding activities of heterogeneous nuclear ribonucleoprotein A3 in vitro and in vivo. Biochim Biophys Acta. 2010;1803:1164–74. doi: 10.1016/j.bbamcr.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 44.Han SP, Friend LR, Carson JH, Korza G, Barbarese E, Maggipinto M, Hatfield JT, Rothnagel JA, Smith R. Differential subcellular distributions and trafficking functions of hnRNP A2/B1 spliceoforms. Traffic. 2010;11:886–98. doi: 10.1111/j.1600-0854.2010.01072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loyer P, Trembley JH, Grenet JA, Busson A, Corlu A, Zhao W, Kocak M, Kidd VJ, Lahti JM. Characterization of cyclin L1 and L2 interactions with CDK11 and splicing factors: influence of cyclin L isoforms on splice site selection. J Biol Chem. 2008;283:7721–32. doi: 10.1074/jbc.M708188200. [DOI] [PubMed] [Google Scholar]
- 46.Yang L, Li N, Wang C, Yu Y, Yuan L, Zhang M, Cao X. Cyclin L2, a novel RNA polymerase II-associated cyclin, is involved in pre-mRNA splicing and induces apoptosis of human hepatocellular carcinoma cells. J Biol Chem. 2004;279:11639–48. doi: 10.1074/jbc.M312895200. [DOI] [PubMed] [Google Scholar]
- 47.Fushimi K, Ray P, Kar A, Wang L, Sutherland LC, Wu JY. Up-regulation of the proapoptotic caspase 2 splicing isoform by a candidate tumor suppressor, RBM5. Proc Natl Acad Sci U S A. 2008;105:15708–13. doi: 10.1073/pnas.0805569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kobayashi T, Ishida J, Musashi M, Ota S, Yoshida T, Shimizu Y, Chuma M, Kawakami H, Asaka M, Tanaka J, Imamura M, Kobayashi M, et al. p53 transactivation is involved in the antiproliferative activity of the putative tumor suppressor RBM5. Int J Cancer. 2011;128:304–18. doi: 10.1002/ijc.25345. [DOI] [PubMed] [Google Scholar]
- 49.Salomonis N, Schlieve CR, Pereira L, et al. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci U S A. 2010;107:10514–9. doi: 10.1073/pnas.0912260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The estrogen receptor α (ER-α) immunoreactivity in the nucleus of epithelial cells in ducts and Lob 1 is similar between parous and nulliparous breasts, as shown in the box plot.