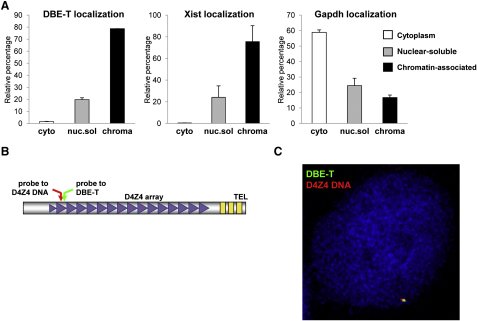
Figure 4.
DBE-T Is Nuclear, Chromatin-Associated, and Localized to the FSHD Locus
(A) Total RNA from AZA+TSA treated chr4/CHO cells was separated into cytoplasmic, nuclear-soluble, and chromatin-bound fractions. The relative abundance of DBE-T in the different fractions was measured by qRT-PCR. As control, Gapdh and Xist were analyzed. The error bars represent SEM.
(B) Schematic representation of the location of the DBE-T and D4Z4 probes.
(C) Following AZA+TSA treatment, chr4/CHO cells were analyzed by RNA/DNA FISH. A single Z stack acquired with an Olympus IX70 DeltaVision RT Deconvolution System microscope is shown. Signals colocalization was detected in 98.8% of the double positive cells (n = 80) deriving from 3 independent experiments. DBE-T is visualized in green, and the D4Z4 DNA is in red. DAPI is in blue.
See also Figures S6 and S7.