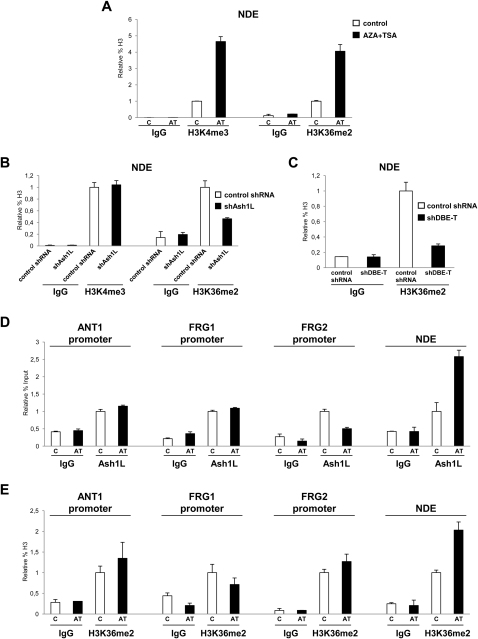
Figure S8.
Ash1L Dimethylates Lysine 36 on Histone H3 at the FSHD Locus, Related to Figure 6
(A) ChIP-qPCR for enrichment on total H3 of H3K4me3, H3K36me2 and IgG, as control, performed on human chr4/CHO cells in the repressed (control) or in the de-repressed (AZA+TSA) state. ChIP material was analyzed by qPCR with primers specific for the NDE region. The error bars represent SEM.
(B) ChIP-qPCR for enrichment on total H3 of H3K4me3, H3K36me2 and IgG, as control, performed in control shRNA or shAsh1L expressing cells in the de-repressed state (AZA+TSA). ChIP material was analyzed by qPCR with primers specific for the NDE region. The error bars represent SEM.
(C) ChIP-qPCR for enrichment on total H3 of H3K36me2 and IgG, as control, in control shRNA or shDBE-T expressing cells in the de-repressed state (AZA+TSA). ChIP material was analyzed by qPCR with primers specific for the NDE region. The error bars represent SEM.
(D) ChIP for Ash1L and IgG, as control, on human chr4/CHO cells in the repressed (control) or de-repressed (AZA+TSA) state. Analysis was performed by qPCR with primers specific for ANT1, FRG1, FRG2 promoters and NDE. Results are expressed as relative percentage of input. The error bars represent SEM.
(E) ChIP-qPCR for enrichment on total H3 of H3K36me2 and IgG, as control, performed on human chr4/CHO cells in the repressed (control) or in the de-repressed (AZA+TSA) state. Analysis was performed by qPCR with primers specific for ANT1, FRG1, FRG2 promoters and NDE. Results are expressed as relative percentage of H3. The error bars represent SEM.