Fig. 4.
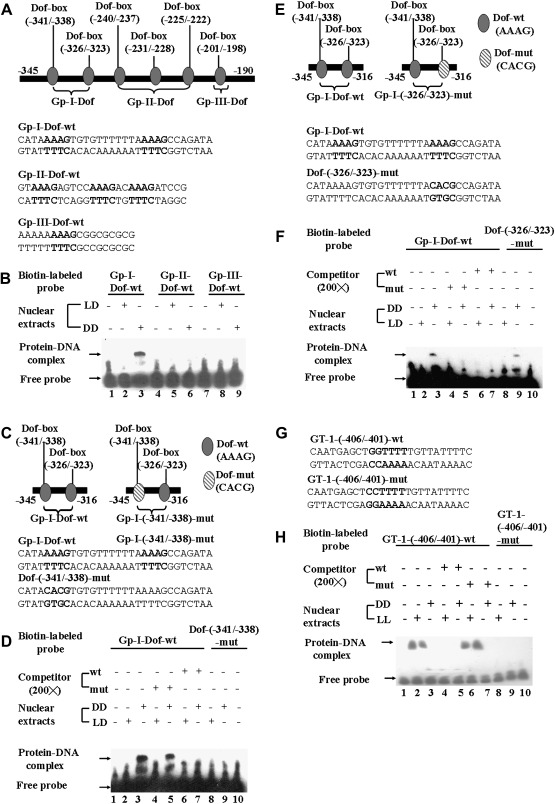
EMSAs on the Dof-box (–341/–338) and GT-1 cis-element (–406/–401) in vitro. (A) Schematic illustration of six putative Dof-boxes on the ACBP3 5'-flanking region. The oval-shaped Dof-boxes were artificially separated into three subgroups, and nucleotide sequences of double-stranded oligonucleotides used in EMSAs are marked below the corresponding Dof groups. The nucleotides of six Dof-boxes are shown in bold. (B) Interaction of nuclear extracts from 3-week-old Arabidopsis leaves with Gp-I-Dof-wt, Gp-II-Dof-wt, and Gp-III-Dof-wt probes, respectively. Crude nuclear extracts (5 μg) from 48 h dark-treated Arabidopsis (DD in lanes 3, 6, and 9) or LD-grown plants (LD in lanes 2, 5, and 8) were incubated with biotin end-labelled Gp-I-Dof-wt (lanes 2 and 3), Gp-II-Dof-wt (lanes 5 and 6), and Gp-III-Dof-wt (lanes 8 and 9) probes. Lanes 1, 4, and 7 are free probes without addition of crude nuclear extracts. (C) Schematic illustration of the mutated Dof-box (–341/–338) (right panel) and its corresponding location in the Gp-I–Dof (left panel) sequence. Nucleotide sequences of double-stranded oligonucleotides used in EMSAs are marked below the corresponding Dof-boxes. Mutated nucleotides in Dof-box (–341/–338) and its corresponding sequences are shown in bold. (D) Interaction of nuclear extracts from 3-week-old Arabidopsis leaves with Gp-I-Dof-wt and Dof-(–341/–338)-mut probes. Crude nuclear extracts (5 μg) from 48 h dark-treated plants (DD in lanes 3, 5, 7, and 9) or control plants (LD in lanes 2, 4, 6, and 8) were incubated with biotin end-labelled Gp-I-Dof-wt (lanes 2–7) or Dof-(–341/–338)-mut (lanes 8 and 9) probes, in the absence (lanes 2, 3, 8, and 9) or presence of a 200-fold molar excess of unlabelled competitor, Gp-I-Dof-wt (lanes 6 and 7), or Dof-(–341/–338)-mut (lanes 4 and 5). Lanes 1 and 10 are free probes without addition of crude nuclear extracts. (E) Schematic illustration of the mutated Dof-box (–326/–323) (right panel) and its corresponding location in the Gp-I–Dof (left panel) sequence. Nucleotide sequences of double-stranded oligonucleotides used in EMSAs are marked below the corresponding Dof-boxes. Mutated nucleotides in Dof-box (–326/–323) and its corresponding sequences are shown in bold. (F) Interaction of nuclear extracts from 3-week-old Arabidopsis leaves with Gp-I-Dof-wt and Dof-(–326/–323)-mut probes. Crude nuclear extracts (5 μg) from 48 h dark-treated plants (DD in lanes 3, 5, 7, and 9) or control plants (LD in lanes 2, 4, 6, and 8) were incubated with biotin end-labelled Gp-I-Dof-wt (lanes 2–7) or Dof-(–326/–323)-mut (lanes 8 and 9) probes, in the absence (lanes 2, 3, 8, and 9) or presence of a 200-fold molar excess of unlabelled competitor, Gp-I-Dof-wt (lanes 6 and 7), or Dof-(–326/–323)-mut (lanes 4 and 5). Lanes 1 and 10 are free probes without addition of crude nuclear extracts. (G) Nucleotide sequences of double-stranded oligonucleotides used in EMSAs for characterization of the GT-1 cis-element in the ACBP3 5'-flanking region. The mutated nucleotides in GT-1-(–406/–401)-mut and their corresponding sequences in GT-1-(–406/–401)-wt are shown in bold. (H) Interaction of nuclear extracts from 48 h dark-treated (DD) or 48 h light-treated (LL) 3-week-old Arabidopsis leaves with GT-1-(–406/–401)-wt and GT-1-(–406/–401)-mut probes. Crude nuclear extracts (5 μg) from dark-treated (DD in lanes 3, 5, 7, and 9) or light-treated plants (LL in lanes 2, 4, 6, and 8) were incubated with biotin end-labelled GT-1-(–406/–401)-wt (lanes 2–7) or GT-1-(–406/–401)-mut (lanes 8 and 9) probes, in the absence (lanes 2, 3, 8, and 9) or presence of a 200-fold molar excess of unlabelled competitor, GT-1-(–406/–401)-wt (lanes 4 and 5), or GT-1-(–406/–401)-mut (lanes 6 and 7). Lanes 1 and 10 are free probes without addition of crude nuclear extracts.