Abstract
In which cells of the flower volatile biosynthesis takes place is unclear. In rose and snapdragon, some enzymes of the volatile phenylpropanoid/benzenoid pathway have been shown to be present in the epidermal cells of petals. It is therefore generally believed that the production of these compounds occurs in these cells. However, whether the entire pathway is active in these cells and whether it is exclusively active in these cells remains to be proven. Cell-specific transcription factors activating these genes will determine in which cells they are expressed. In petunia, the transcription factor EMISSION OF BENZENOIDS II (EOBII) activates the ODORANT1 (ODO1) promoter and the promoter of the biosynthetic gene isoeugenol synthase (IGS). The regulator ODO1 in turn activates the promoter of the shikimate gene 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS). Here the identification of a new target gene of ODO1, encoding an ABC transporter localized on the plasma membrane, PhABCG1, which is co-expressed with ODO1, is described. PhABCG1 expression is up-regulated in petals overexpressing ODO1 through activation of the PhABCG1 promoter. Interestingly, the ODO1, PhABCG1, and IGS promoters were active in petunia protoplasts originating from both epidermal and mesophyll cell layers of the petal, suggesting that the volatile phenylpropanoid/benzenoid pathway in petunia is active in these different cell types. Since volatile release occurs from epidermal cells, trafficking of (volatile) compounds between cell layers must be involved, but the exact function of PhABCG1 remains to be resolved.
Keywords: ABC transporter, cell specificity, floral fragrance, petunia, transcription factor
Introduction
The reproductive success of many flowering plants depends on scent production by their flowers, but the biosynthetic pathways that produce the precursors of these volatile compounds are involved in more processes. For instance, in addition to their role in floral scents, phenylpropanoids function in UV protection, lignification, protection against pathogens and herbivores, and flower pigmentation (Vogt, 2010). In addition, the compounds these pathways produce are used in the flavour and fragrance industry, and manipulation of these pathways could be used to improve floral fragrance (Dudareva and Pichersky, 2008) as well as fruit flavour. Petunia hybrida has become one of the models of choice to study floral volatile biosynthesis and regulation, and its floral headspace is dominated by volatile benzenoids and phenylpropanoids, which are produced from shikimate-derived L-phenylalanine (Boatright et al., 2004). Several genes encoding shikimate enzymes (Colquhoun et al., 2010a; Maeda et al., 2010), and the subsequent branched pathways leading to the production of C6–C3 (Koeduka et al., 2006; Dexter et al., 2007), C6–C2 (Kaminaga et al., 2006), and C6–C1 (Negre et al., 2003; Boatright et al., 2004; Van Moerkercke et al., 2009) volatile compounds, have been identified and characterized in petunia. The transcriptional network underlying scent production in flowers is partially understood, and knowledge about the cell specificity of floral fragrance production is limited.
The petunia R2R3-MYB transcription factor (TF) ODORANT1 (ODO1), a regulator of floral fragrance production, is co-expressed with several genes of the volatile benzenoid/phenylpropanoid pathway, and was shown to activate the promoter of the shikimate gene 5-enolpyruvylshikimate-3-phospate synthase (EPSPS), thereby regulating precursor supply for L-phenylalnine and volatile production (Verdonk et al., 2005). The R2R3-MYB TF EMISSION OF BENZENOIDSII (EOBII) activates the promoters of petunia isoeugenol synthase (IGS) and the tobacco PALB (Spitzer-Rimon et al., 2010). However, silencing of these two regulators in petunia flowers affected transcript levels of many more genes (Verdonk et al., 2005; Spitzer-Rimon et al., 2010), which are thus potential target genes of these TFs. Subsequent promoter studies identified EOBII as a direct activator of ODO1 (Van Moerkercke et al., 2011).
Whereas the biochemistry and molecular biology of floral scent is well understood, the cell biology has remained understudied. However, the involvement of plastids, peroxisomes, and mitochondria was recently shown (Long et al., 2009; Van Moerkercke et al., 2009; Maeda et al., 2010). After biosynthesis, volatile compounds need to be trafficked across the cell membrane, the cell wall, and the cuticle layer, before they are emitted (Jetter, 2006). In addition, trafficking between cell layers might be needed if different cell layers of the petal are involved in the production of these compounds. Whether this trafficking involves diffusion, active transport, or both is uncertain (Jetter, 2006), but ATP-binding cassette (ABC) transporters could facilitate transport of compounds across membranes. For instance, the Arabidopsis ABCG11 (WBC11/DSO/COF) (Bird et al., 2007; Panikashvili et al., 2007), ABCG12 (WBC12/CER5) (Pighin et al., 2004), and ABCG13 (WBC13) (Panikashvili et al., 2011) are involved in cutin and/or wax deposition at the surface of Arabidopsis organs. Also, the Nicotiana plumbaginifolia ABC1 (NpPDR1) actively secretes the terpenoid sclareol from sclareol-treated cells (Jasinski et al., 2001). ATP-dependent transport of aglycone monolignols across plasma membrane vesicles was demonstrated recently (Miao and Liu, 2010). Importantly, some of the monolignols tested, notably coniferylalcohol and coniferaldehyde, are precursors for the C6–C3 volatile phenylpropanoids eugenol and isoeugenol (Koeduka et al., 2006; Dexter et al., 2007), which are also emitted by petunia flowers.
Like volatile phenylpropanoids/benzenoids, anthocyanins originate from the shikimate pathway, and their production, as well as cellular localization and regulation of the genes involved, is well defined (Koes et al., 2005). Transcripts of late anthocyanin biosynthetic genes, such as dihydroflavonol-4-reductase a (DFRa), and their regulators were shown to act exclusively in the epidermal cells of petunia petals (Quattrocchio et al., 1998, 2006; Faraco et al., 2011) and, accordingly, anthocyanins exclusively accumulate in these cells (Koes et al., 1990). Recently, it was shown that protoplasts originating from petal limbs of Petunia hybrida M1×V30 retain cell-specific promoter activity of genes, as was illustrated for the promoter of DFRa (Faraco et al., 2011). The cell type specificity of the shikimate pathway in petunia petals is not entirely clear. The EPSPS promoter shows activity in the epidermal cell layer in petals of flower buds of the white P. hybrida cv. W80. In contrast, high activity in mature flowers is seen in both epidermis and mesophyll layers of the petal (Benfey et al., 1990). Since the production of floral pigments occurs before anthesis, it is not likely that expression of EPSPS at later stages of flower development is related to pigment biosynthesis, but is more likely to be related to volatile biosynthesis. Therefore, EPSPS promoter activity in the mesophyll layer raises questions about the cell type specificity of fragrance biosynthesis in petunia flowers. Here, the cell-specific activity of several genes was determined in order to elucidate the spatial organization of floral scent production.
Materials and methods
Plant material and protoplast isolation
Petunia×hybrida cv. Mitchell (also referred to as cv. W115) and cv. R27, Petunia×hybrida M1×V30, and Nicotiana benthamiana were grown in standard greenhouse conditions (16 h photoperiod, 300–400 μmol m−2 s−1 light intensity, 60–65% humidity, and day/night temperatures of 22/17 °C). Plants were moved to controlled growth chambers (16 h photoperiod, 250–350 μmol m−2 s−1 light intensity, 70% relative humidity, and constant temperature of 21 °C) at least 3 d prior to the experiments, except where otherwise stated. For protoplast preparation, M1×V30 plants were moved to the laboratory 2 d prior to isolation. Protoplasts were prepared as described by Faraco et al. (2011).
Construct design
For the PhABCG1 promoter–reporter construct [ABCG1:GUS (β-glucuronidase)], a 2005 bp fragment of the promoter including the 5′-untranslated region (UTR) was PCR amplified introducing an AscI site at the 5′ end and an NcoI site at the ATG, and cloned via AscI/NcoI upstream of an intron-containing the uidA CDS (GUS) at the start codon followed by the Nos terminator. The pABCG1:GUS-tNos cassette was subsequently cloned via AscI/EheI into pBINPLUS (Vanengelen et al., 1995) pre-digested with AscI and SmaI. For the 35S-driven green fluorescent protein (GFP)–ODO1 construct (35S:GFP–ODO1), GFP was PCR amplified, introducing an NcoI site at the ATG and an alanine–glycine linker at the 3′ end with an XbaI site excluding the stop codon (fw primer, 5′-GTGAACCATGGTGAGCAAGGGCGAGGAG-3′; rev primer, 5′-GGTCTAGAACCAGCCTTGTACAGCTCGTCCATG-3′; restriction sites are underlined; the alanine–glycine linker is in bold). The ODO1 CDS was PCR amplified, introducing an XbaI site followed by an alanine–glycine linker preceding the ATG and an SacI site immediately after the stop codon (fw primer, 5′-GGTCTAGAGCTGGTATGGGAAGACAACCT TG-3′; rev primer, 5′-ATGAGCTCTTATCCGTGCCTGTTCTCTAC-3′; restriction sites are underlined; the alanine–glycine linker is in bold; the start and stop codon are in italics). Using a three-point ligation, the GFP and ODO1 fragments were ligated via NcoI–SacI in a shuttle vector between the Cauliflower mosaic virus (CaMV) 35S promoter and the nopaline synthase terminator (tnos). The 35S:GFP–ODO1:tnos cassette was then excised using HindIII and SfoI and ligated via HindIII–SmaI into pBINPLUS (Vanengelen et al., 1995). To create the ODO1:GFP–ODO1 construct, a 1.9 kbp fragment of the ODO1 promoter was first PCR amplified introducing a HindIII site at the 5′ end and an XbaI site preceding the start codon (fw primer, 5′-CCCAAGCTTGAAGGGGGATCTCATG-3′ and rev primer, 5′- GGGGTACCAACGACTTAACGACAATAG-3′) and exchanged with the 35S promoter in the shuttle vector described above, which has an XbaI site immediately upstream of the NcoI site. The construction of 35S:ODO1, 35S:EOBII, 35S:AN2, and 35:LUC has been described previously (Quattrocchio et al., 1999; Verdonk, 2006; Van Moerkercke et al., 2011). The 35S-driven mVenus-PhABCG1 construct (35S:mVenus-ABCG1) was created using multisite Gateway technology (Invitrogen). The PhABCG1 genomic region including the stop codon was PCR amplified, introducing the AttB recombination sites with primers 5′- AAAAAGCAGGCTCTATGACAAACCAATTACCAAAC-3′ and 5′- AGAAAGCTGGGTACTAGGAATCAAATACATCCA-3′ (stop codon is in italics) and subsequently recombined into pDONR201 (BP reaction; Invitrogen), creating PhABCG1-pENTR, which was then recombined into a destination vector (LR reaction), containing the CaMV 35S promoter, along with the entry clones pGEM Box1 mVenus (Nagai et al., 2002) and pGEM Box3 tNos. The ABCG1:mVenus-ABCG1 construct was created by replacing the CaMV 35S promoter in the 35S:mVenus-ABCG1 construct with a 2 kb PhABCG1 promoter fragment of P. hybrida cv. Mitchell. For this, the promoter was first PCR amplified introducing SalI and Acc65I sites (fw primer, 5′-CCCGTCGACTATTAGTATACTGTTCAATTC-3′; rev primer, 5′-GGGGTACCCAACGACTTAACGACAATAG-3′; the respective restriction sites are underlined) and the fragment was subsequently cloned via XhoI/Acc65I into the pre-digested 35S:mVenus-ABCG1 destination vector.
Expression analyses
For RNA gel blot analyses, 2-day-old flowers of P. hybrida cv. Mitchell plants were used and independent sample collections for all experiments were performed twice. For the time course, 2-day-old wild-type petals were collected during a 30 h period with 3 h intervals. For the developmental course, wild-type petal tissue of flowers in various stages of development (1 cm bud, 2 cm bud, 3 cm bud, 4 cm bud, flower opening, 1-day-old open flower, 3-day-old open flower, and senescent flower) was collected 2 h before the onset of the dark period. The different tissues were collected from wild-type plants 2 h before the onset of the dark period. Total RNA was extracted using TriZol (Invitrogen). An 8 μg aliquot was loaded on a gel and the blots were hybridized using a PhABCG1-specific probe (forward primer, 5′-CTCAATTCATCAACCTAG TGTTG-3′; reverse primer, 5′-GGTATTTCCAAAATGGCTTAGG-3′). To visualize loading of the gel, the tissue blot was re-hybridized with an 18S probe (forward primer, 5′-AGCAGGCTAAGGTCTCGT-3′; reverse primer, 5′-AGCGGATGT TGCTTTTAGGA-3′). For the developmental course and time course a petunia floral binding protein 1 (FBP1) probe was used (forward primer, 5′-GTGTTCTTTGTGATGCTCGTG-3′; reverse primer, 5′-CAACCTCTCCTGCAAATTTGG-3′) to show loading of the gel (Verdonk et al., 2005; Van Moerkercke et al., 2009). Isolation of petal RNA and hybridization using ODO1 and FBP1 probes for the ODO1-silenced stable lines was described by Verdonk et al. (2005), and the blot was re-hybridized with a PhABCG1 probe.
For RT-PCR, wild-type petals of P. hybrida cv. Mitchell and P. hybrida cv. R27 were collected in winter in the greenhouse 1 h before artificial illumination ceased, when it was dark outside, and volatile emission was high. For the transient ODO1 overexpression experiment, petals of Mitchell were infiltrated with Agrobacterium tumefaciens GV3101 (pMP90) harbouring either a 35S:LUC or a 35S:ODO1 construct, incubated for 36 h, and sampled 12 h before the onset of the artificial dark period in the growth chamber when volatile emission was low. Total RNA was extracted using the Qiagen Plant RNA extraction kit and 1 μg was used for cDNA synthesis in a 20 μl reaction volume after DNase (Invitrogen) treatment. Semi-quantitative RT-PCR was performed in a 25 μl reaction using 1 μl of 10× diluted cDNA samples with a standard PCR program and Taq polymerase (DreamTaq, Fermentas). Twenty cycles were performed using forward primer 5′-CTCAATTCATCAACCTAGTGTTG-3′ and reverse primer 5′-GGTATTTCCAAAATGGCTTAGG-3′ for PhABCG1, and primers 5′-GTTGGTGGTAGCTGAGAGTCAG-3′ and 5′-GACTCTAAGCAAATCTAACTTCC-3′ for ODO1. Amplification of FBP1 (22 cycles) was used to show that approximately equal amounts of input cDNA were used, employing forward primer 5′-GTGTTCTTTGTGATGCTCGTG-3′ and reverse primer 5′-CAACCTCTCCTGCAAATTTGG-3′.
For quantitative RT-PCR, forward primer 5′-GTTATCCTTTTCATATAAGC-3′ and reverse primer 5′-CTCAACAAATATGCATCGCATAC-3′ were used to amplify PhABCG1. ACTIN (forward primer 5′-TGCTGATCGTATGAGCAAGGAA-3′ and reverse primer 5′-GGTGGAGCAACAACCTTAATCTTC-3′; Spitzer-Rimon et al., 2010) was used to enable normalization. Analysis was performed as described by Van Moerkercke et al. (2011). Two technical and three biological replicates were used.
Isolation of the cDNA, the promoter, and the genomic region of PhABCG1
The PhABCG1 clone was identified from a cDNA library, of which the first strand was generated from the poly(A) tail. The full-length cDNA was then isolated using a 5′-RACE (rapid amplification of cDNA ends) protocol (Invitrogen). The genomic region was PCR isolated using primers located in the 5′-UTR and 3′-UTR of the gene. The promoter region of PhABCG1 was isolated using a genome walking protocol (Siebert et al., 1995) as described by Van Moerkercke et al. (2011), using PhABCG1-specific primers.
Promoter analyses and trans-activation assays using agro-infiltration
For promoter analysis in petal limbs and the trans-activation assays, the ABCG1:GUS reporter construct was used. GUS activity in petals was determined as described in Van Moerkercke et al. (2011). For the trans-activation assays, a 35S:ODO1 was used as the effector construct. As a control, 35S:RFP, 35S:AN2, or 35S:EOBII were used. Trans-activation assays in petunia petals and N. benthamiana leaves were performed as described by Van Moerkercke et al. (2011).
Confocal laser scanning microscopy
All imaging was performed using a ×20 water objective with a Nikon Eclipse T1 microscope coupled to a Nikon A1 confocal scanning head. In P. hybrida cv. Mitchell petals (Fig. 6), mVenus fluorescence was monitored with a 525–555 nm BP emission filter (488 nm excitation); FM4-64 (Molecular Probes, Invitrogen) was visualized with a 570–620 nm BP emission filter (488 nm excitation). In N. benthamiana leaves (Fig. 7), GFP (488 nm excitation), mCherry (561 nm excitation), and autofluorescence (488 nm excitation) were monitored using 550–550 nm BP, 570–620 nm BP, and 605 nm LP emission filters, respectively. For visualization of fluorescence in protoplasts in Fig. 8, mTurquoise (443 nm excitation), GFP and mVenus (514 nm excitation), and autofluorescence (443 nm excitation) were monitored with 464–499 nm BP, 525–555 nm BP, and 605 nm LP emission filters, respectively. The 35S-driven mTurquoise membrane marker was created in pMON999 by fusing the hypervariable region of maize ROP7 (Ivanchenko et al., 2000) to mTurquoise (Goedhart et al., 2010). For visualization of fluorescence in protoplasts in Fig. 9, excitation was provided by 457 nm and 561 nm lasers. To monitor mTurquoise, dsRed/autofluorescence, and autofluorescence, the 464–499 nm BP, 570–620 nm band pass, and 662–737 nm BP emission filters, respectively, were used. The images were processed by ImageJ (http://rsb.info.nih.gov/ij/) and assembled in MS Power Point.
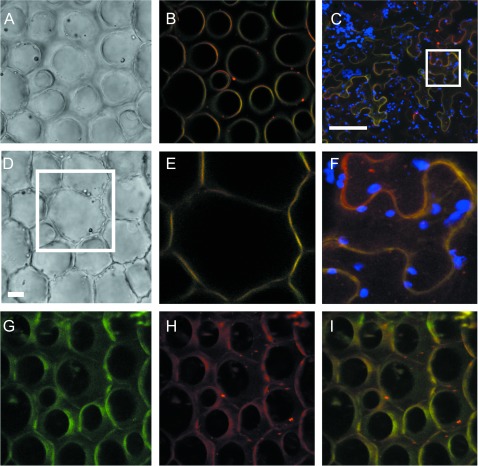
Fig. 6.
PhABCG1 localizes to the plasma membrane. (A, D) Confocal images of petunia petal cells after agroinfiltration with 35S:ABCG1-mVenus and incubation with the plasma membrane marker FM4-64, showing the conical epidermal cells near the tip (A) and at the base (D). (B) Single confocal section of the image in A, showing the merged signal (yellow) of mVenus (green) and FM4-64 (red). (E) Single confocal section of the boxed area in D showing the merged image of the mVenus and FM4-64 signals at the cell base. The scale bar in D represents 10 μm. (G–I) Confocal projection images of the conical epidermal cells in A and D for mVenus (G) and FM4-64 (H) showing co-localization at the plasma membrane (I). The images are three-dimensional reconstructions of multiple confocal images, including the one depicted in B, shown at an angle of 45 ° to illustrate co-localization of the signals at the surface of the conical epidermal cells. (C) Merged image of a single confocal section of infiltrated epidermal cells of N. benthamiana leaves showing mVenus–PhABCG1 (green), FM4-64 (red), and autofluorescence (blue). (F) Magnification taken from the boxed area in C. The scale bar in C represents 100 μm.
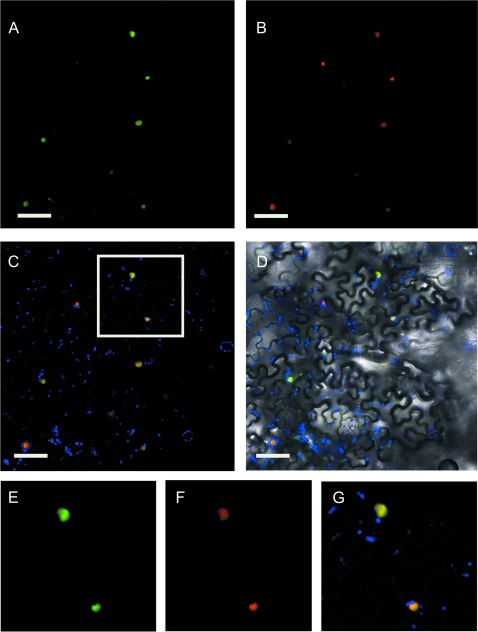
Fig. 7.
Intracellular localization of ODO1. (A, B) Confocal microscopy images showing subcellular localization of 35S:GFP–ODO1 (A) and 35S:NLS–mCherry (B) in agro-infiltrated leaves of Nicotiana benthamiana. Co-localization at the nucleus (yellow) of GFP (green) and mCherry (red) is shown (C). Chloroplast autofluorescence is shown in blue (C, D) and the corresponding transmitted light image in D. (E–G) Digital magnification from the boxed area in C showing GFP–ODO1 (green) (E), NLS–mCherry (red) (F), and the corresponding merged image (G). The scale bar represents 20 μm.
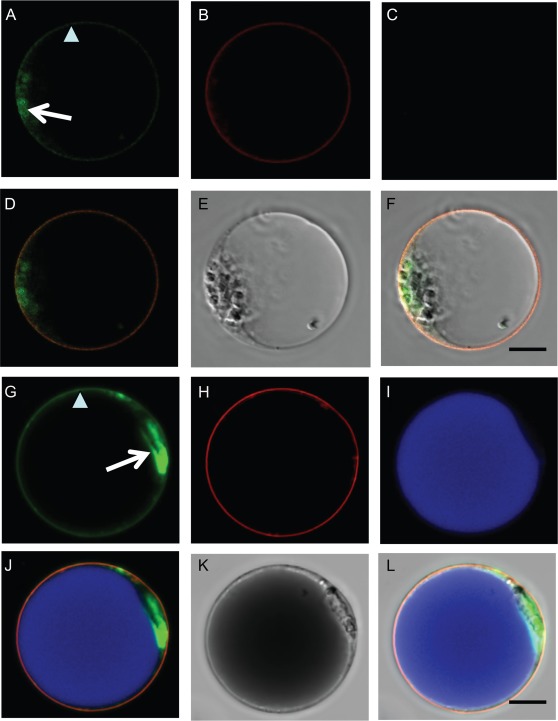
Fig. 8.
Cell type-specific activation of the ODO1 and PhABCG1 promoters in M1×V30 petunia petal protoplasts. Protoplasts derived from mesophyll (A–F) and epidermal (G–L) cells of the petal were co-transfected with ODO1:GFP–ODO1 and ABCG1:mVenus–ABCG1. (A, G) ODO1 nuclear (arrow) and PhABCG1 plasma membrane (arrowhead) subcellular localizations were detected simultaneously (green). (B, H) Expression of a 35S-driven mTurquoise plasma membrane marker (red). (C, I) Autofluorescence of anthocyanins in the vacuole of epidermal-derived protoplasts in I (blue). Corresponding merged images (D, J) showing GFP and mVenus (green), the mTurquoise membrane marker (red), and autofluorescence (blue). Bright field (E) and merged image of A–E (F) of a mesophyll cell-derived protoplast. Bright field (K) and merged image of G–K (L) of an epidermal cell-derived protoplast. The scale bar represents 10 μm.
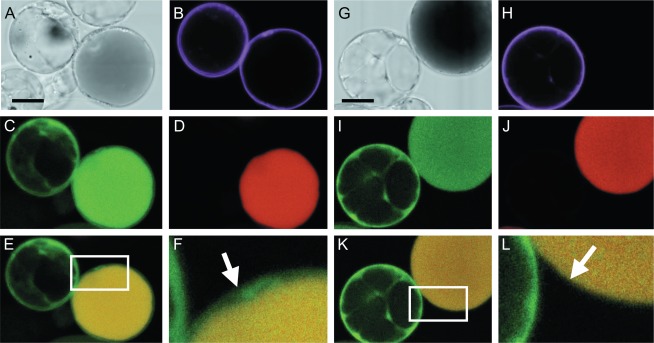
Fig. 9.
Activation of the petunia IGS promoter in petunia M1×V30 protoplasts. (A) Bright field image of the protoplast shown in B–E. (B) Transformation control showing expression of an mTurquoise membrane marker (magenta) in a protoplast derived from the epidermis (right) and a protoplast derived from the mesophyll (left; see D). (C) Green signal from expression of IGS:dsRed and/or autofluorescence of the anthocyanins in the vacuole. (D) Far-red signal from autofluorescence of anthocyanins in the vacuole (red). (E) Merged image of the single confocal sections depicted in C and D showing autofluorescence in the vacuole (yellow) and dsRed signal (green) from IGS:dsRed in the cytosol. (F) Magnification of the boxed area in E. The arrow shows the dsRed signal (green) in the cytosol of an epidermal protoplast. (G–J) Similar to A–D, but the epidermal-derived protoplast is not transformed (H). (K) dsRed signal from IGS:dsRed (green) in the mesophyll protoplast. (L) Magnification of the boxed area in K. The arrow shows the absence of dsRed signal (green) in the non-transformed epidermal protoplast. The scale bars represent 10 μm.
Accession numbers
The sequence corresponding to the PhABCG1 genomic region has been deposited in GenBank (accession no. JQ088099).
Results
Identification of an ODORANT1-dependent gene encoding a putative half-size ATP-binding cassette transporter of class G
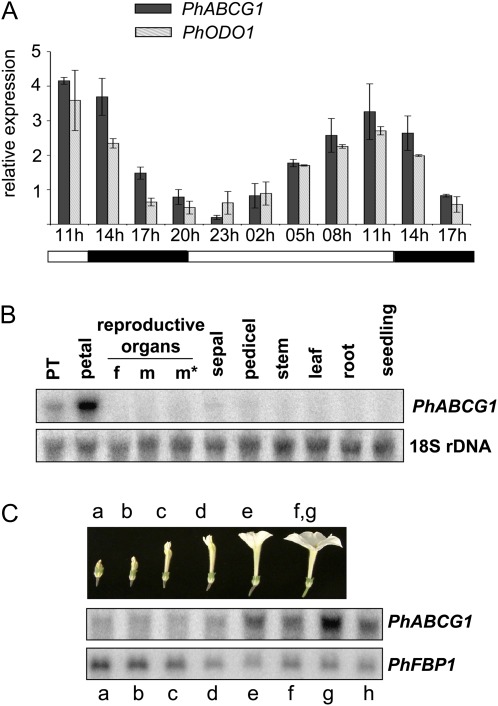
Microarray analysis performed with RNA extracted from petal limbs of P. hybrida cv. Mitchell, which was collected at two different time points during the day/night cycle (Verdonk et al., 2003; Verdonk, 2006), revealed a partial cDNA encoding a putative ABC protein of subclass G, which was named P. hybrida ABCG1. PhABCG1 was highly up-regulated 3 h before the peak of scent emission, but not when emission was low (26-fold induction, P=0.006, n=3). Three hour interval sampling of Mitchell floral petals and subsequent RNA blot analysis confirmed rhythmic transcript accumulation for this gene during the day and night, which occurred in-phase with ODO1 expression (Fig. 1A). Further analysis showed the strongest expression in the petal limbs and to a lesser extent in petal tubes, whereas expression in any other tissue could barely be detected (Fig. 1B). Transcript abundance increased during flower development and peaked in open flowers, and dropped as flowers senesced (Fig. 1C). This combined expression pattern is similar to and specific for genes of the volatile benzenoid/phenylpropanoid pathway in petunia and is thus indicative of a role in floral scent production (Schuurink et al., 2006).
Fig. 1.
Characterization of PhABCG1 expression. RNA gel blots were hybridized with the indicated probes. Re-hybridization with a FLORAL BINDING PROTEIN 1 (FBP1) or an 18S rDNA probe was used to show loading of the gel or to enable normalization. (A) Rhythmic expression of PhABCG1 in Mitchell flowers, in phase with ODO1 expression and rhythmic volatile emission (not shown; Verdonk et al., 2003). Mean values are shown (n=2). Error bars indicate maximum and minimum values. Black and grey bars indicate PhABCG1 and ODO1 transcript levels, respectively. Petal tissue was harvested at the time points indicated below the graph (3 h intervals). Black and white bars below the graph indicate dark and light conditions, respectively. (B) Tissue-specific accumulation in petunia Mitchell. The tissues used are indicated (f, style and stigma; m, non-dehisced anthers; m*, dehisced anthers; PT, petal tube; petal refers to the petal limb). (C) Developmentally regulated transcript accumulation in Mitchell petals. Developmental stages of the petals: a–e, as indicated; f and g indicate open flowers before and after anthesis, respectively; h indicates a senescing flower.
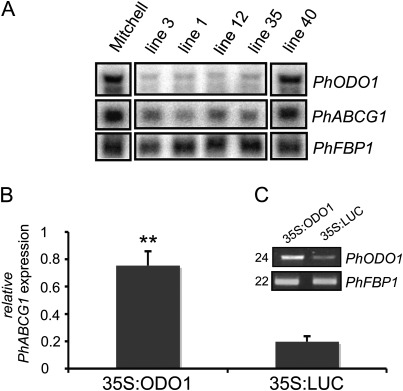
Transcript analysis using the previously established petunia ODO1 RNA interference (RNAi) knock-down lines (Verdonk et al., 2005) showed reduced PhABCG1 transcript accumulation in those lines that were successfully silenced, suggesting a dependence on ODO1 expression (Fig. 2A). To corroborate this, ODO1 was transiently overexpressed in Mitchell petals and PhABCG1 expression levels were subsequently monitored using quantitative RT-PCR at a time point when ODO1 and PhABCG1 expression is normally low (Fig. 1A). Figure 2C shows increased ODO1 transcript levels, resulting in 4-fold higher PhABCG1 levels (Fig. 2B), compared with a 35S:LUCIFERASE control infiltration (P < 0.001, n=3).
Fig. 2.
Expression of ODO1 influences PhABCG1 transcript levels. (A) RNA gel blot analysis of ODO1-RNAi lines showing reduced levels of PhABCG1 expression. FBP1 was used to show loading of the gel. Line 40 is a transgenic line in which ODO1 is not silenced (Verdonk et al., 2005). (B) Quantitative RT-PCR showing PhABCG1 transcript levels in petals of Mitchell after agro-infiltration with 35S:ODO1 or 35S:LUC at a time point when endogenous transcript levels are low. The mean and SD of three independent biological replicates are shown. Asterisks indicate significant differences (t-test, P < 0.001, n=3). Actin was used as a reference gene and values are relative to Actin. (C) RT-PCR showing transcript levels of ODO1 in the samples in B. The number of cycles used is shown at the left and the floral binding protein 1 (PhFBP1) of petunia was used as a reference gene. A representative gel is shown.
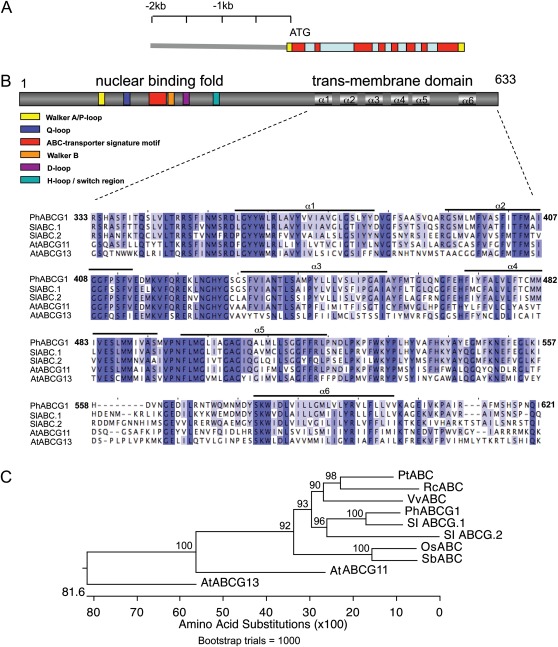
A full-length PhABCG1 transcript was obtained using 5′-RACE. The cDNA was 2178 bp long, with a 146 bp 5′-UTR and 130 bp 3′-UTR. Isolation of the genomic region showed a gene structure with eight exons and seven introns (Fig. 3A). The cDNA putatively encodes a protein of 633 amino acids and the protein contains the highly conserved Walker A and B boxes (Walker et al., 1982), separated by the Q-loop and ABC signature (Higgins et al., 1986) and flanked by the D-loop and H-loop/switch regions, which together correspond to the nuclear binding fold (NBF; Fig. 3B). A Kyte–Doolittle (Kyte and Doolittle, 1982) plot revealed six domains with a high content of hydrophobic residues located at the C-terminus of the polypeptide, corresponding to six putative α-helices (α1–α6 in Fig. 3B) spanning the membrane (transmembrane domain; TMD). In this order and number, the NBF-TMD structure is typical for proteins belonging to the ABCG/WBC subfamily of ABC proteins (Verrier et al., 2008). Homologue proteins were searched for using the BLAST tool of NCBI. The closest characterized protein is the Arabidopsis ABCG11/WBC11/COF1/DSO (Pighin et al., 2004) with 47% amino acid identity between the TMDs. Homology with the floral-specific AtABCG13 (Panikashvili et al., 2011) is 40%. The closest uncharacterized putative homologues are from Ricinus communis (XM002525791; 62% homology between the TMDs), Populus trichocarpa (XM002300152; 61.5%), Vitis vinifera (XM002270611; 62%), Sorghum bicolor (XM002460339; 60%), and Oryza sativa (NM001069981; 60%). A putative homologue ABC transporter was further searched for in the recently sequenced genome of tomato, and two proteins were found that show 78% (SlABC.1) and 57% (SlABC.2) amino acid identity with PhABCG1 between their TMDs (tomato loci Solyc01g105400.2.1 and Solyc01g105450.2.1, respectively; www.solgenomics.net). An alignment of the TMDs of PhABCG1, AtABCG11 and G13, and SlABC.1 and 2, along with a phylogenetic tree of the TMDs of the aforementioned species is shown in Fig. 3B and C, respectively.
Fig. 3.
Structure of PhABCG1 and the encoded protein. (A) The genomic region consists of eight exons (red) and seven introns (blue). The UTRs are in yellow. The 2 kb promoter region used in this study is shown. (B) PhABCG1 encodes a half size ABC transporter of 663 amino acids. The nuclear binding fold and transmembrane domain (TMD) are located at the N-terminus and C-terminus, respectively. Characteristic features in the nuclear binding fold are indicated as boxes. An alignment of the putative TMD with those of the closest homologues in Solanum lycopersicum (SlABC.1, Solyc01g105400.2.1; and SlABC.2, Solyc01g105450.2.1) and with those of Arabidopsis ABCG11 (NM_101647.4) and G13 (NP_175557.1) is shown. Six putative α-helices spanning the membrane are indicated above the amino acid sequence. (C) Phylogenetic tree of a selection of the TMDs used for the alignment in B, extended with those of the closest homologues in Oryza sativa (Os, NM001069981), Populus trichocarpa (Pt, XM002300152), Ricinus communis (Rc, XM002525791), Vitis vinifera (Vv, XM002270611), and Sorghum bicolor (Sb, XM002460339). The alignment was made using CLUSTALW and the tree was constructed according to the Neighbor–Joining method after bootstrap analysis (×1000). Bootstrap values are shown.
ODORANT1 activates the PhABCG1 promoter
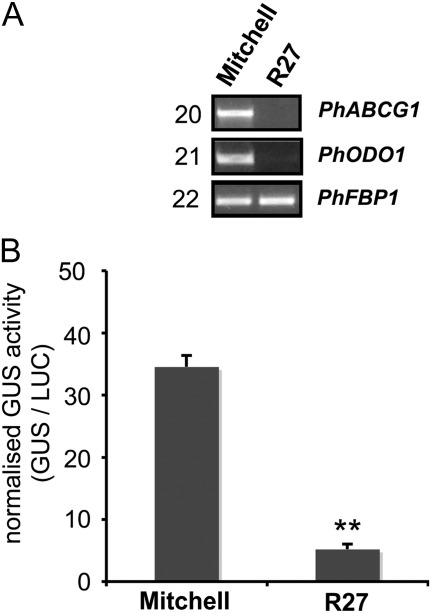
Previously, it has been shown that ODO1 is not expressed in petals of the non-fragrant P. hybrida cv. R27 (Van Moerkercke et al., 2011). Therefore, if PhABCG1 expression depends on ODO1, PhABCG1 expression should be low in petals of R27. Indeed, semi-quantitative RT-PCR analysis could not detect PhABCG1 transcripts in petals of R27 (Fig. 4A). To investigate the role of cis-elements and trans-factors in this expression pattern, the activity of the PhABCG1 promoter from Mitchell was analysed in petal limbs of Mitchell and R27. For this, the 2 kb promoter region (including the 5′-UTR) was fused to an intron-containing β-glucuronidase (uidA or GUS) CDS and the activity in petal limbs of these petunias was assessed using agro-infiltration (Fig. 4B) as described before (Van Moerkercke et al., 2011). The Mitchell promoter was significantly more active in Mitchell petal limbs compared with R27 petal limbs (7-fold, P < 0.005, n=6), meaning that a crucial activator of PhABCG1 is lacking in R27 petals.
Fig. 4.
PhABCG1 promoter activity is correlated with expression of ODO1. (A) RT-PCR showing expression of PhABCG1 and ODO1 in petals of P. hybrida cv. Mitchell and R27. The number of cycles used is indicated on the left. (B) Mitchell PhABCG1 promoter activity in petals of Mitchell and R27. Petals were co-infiltrated with A. tumefaciens harbouring ABCG1:GUS and 35S:LUC (average ±SE, n=4). The experiment was performed twice and the result of a representative experiment is shown. Asterisks indicate significant differences (t-test, P < 0.005, n=6).
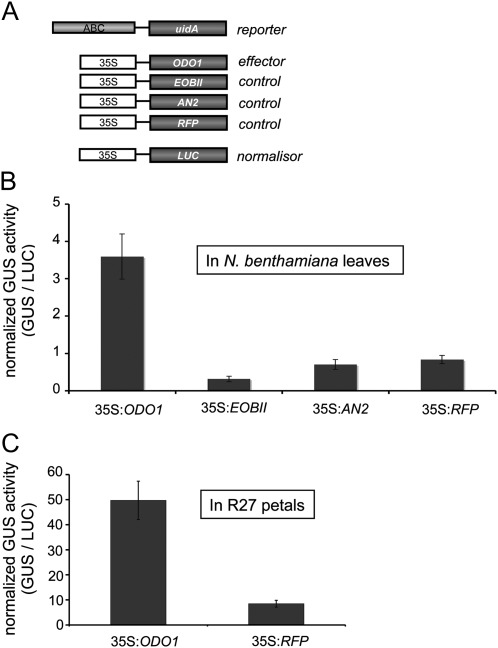
To verify whether ODO1 activates the PhABCG1 promoter, a trans-activation assay in N. benthamiana leaves was used (Fig. 5A) (Lee and Yang, 2006). For this, the ABCG1:GUS reporter construct was expressed together with the effector construct 35S:ODO1. As a control, the same reporter construct was separately co-infiltrated with the R2R3-MYB TFs EOBII, ANTHOCYANIN 2 (AN2), or red fluorescent protein (RFP) effector/control constructs (Fig. 5A). Activity was induced up to 7-fold when co-infiltration with ODO1 was compared with the control infiltrations (RFP; Fig. 5B). Neither of the other two MYB TFs induced expression of the reporter gene. These results indicate that ODO1 activates the PhABCG1 promoter in the leaves of N. benthamiana. Since R27 flowers do not express ODO1 (Fig. 4A) and because the Mitchell promoter is not active in R27 petals (Fig. 4B), PhABCG1 promoter activation by ODO1 was assessed in petals of this petunia as well (Fig. 5C). The 35S:RFP was used as a control infiltration as both AN2 and EOBII are expressed in R27 flowers (Quattrocchio et al., 1993; Van Moerkercke et al., 2011). Similar to leaves, the PhABCG1 promoter could be activated by ectopically expressing ODO1 in R27 petals (Fig. 5C), confirming the identification of ODO1 as a direct activator of PhABCG1 in petunia petal limbs.
Fig. 5.
The PhABCG1 promoter is activated by ODORANT1. (A) Constructs used for the trans-activation assays. (B, C) Trans-activation assay with ectopically expressed effector and control constructs in leaves of N. benthamiana (B) and petals of R27 (C). Co-infiltration with 35S:LUC was used to normalize for infiltration and protein extraction efficiency.
Subcellular localization of PhABCG1 and ODORANT1
To determine the subcellular localization of PhABCG1, an N-terminal translational fusion with the fluorescent protein mVenus was created (Nagai et al., 2002) under control of a double CaMV 35S promoter and it was transiently expressed in N. benthamiana leaves and P. hybrida cv. Mitchell petals using agro-infiltration (Verweij et al., 2008; Van Moerkercke et al., 2009). Initial attempts to create the construct with the PhABCG1 CDS failed, so the full genomic region was used. Confocal imaging of the fusion protein showed co-localization with the cell membrane marker FM4-64 (Bolte et al., 2004) in the conical cells of the adaxial epidermis of petunia petals. Figure 6A and D shows the transmitted light through the adaxial conical cells and at the base of the cells, respectively. Single confocal sections show co-localization of mVenus–ABCG1 with the FM4-64 marker in these cells (Fig. 6B, E). A three-dimensional reconstruction of multiple confocal projection images of the same cells, represented at an angle of 45 °, shows co-localization at the entire surface of these cells (Fig. 6G–I). Co-localization with FM4-64 was also observed in epidermal cells of tobacco leaves (Fig. 6C, F), confirming plasma membrane localization in non-petal tissue.
Although nuclear localization of ODO1 can be predicted from its function as a TF (Verdonk et al., 2005), the protein has not been localized in planta yet. Therefore, N-terminal (35S:GFP–ODO1) and C-terminal (35S:ODO1–GFP) trans- lational fusions to the GFP CDS under control of a CaMV 35S promoter were first created and transiently expressed in leaves of N. benthamiana. As a control, a 35S-driven mCherry CDS containing a nuclear localization signal (NLS) was co-infiltrated. The GFP signal of the C-terminal fusion protein could not be detected by either epifluorescence microscopy or western blot analysis (data not shown), but the GFP signal of 35S:GFP–ODO1 (Fig. 7A) co-localized with the mCherry signal (Fig. 7B) in the nuclei of transformed N. benthamiana leaves (Fig. 7C–G).
ODORANT1 and PhABCG1 expression in petal protoplasts
To investigate the cell layer in which PhABCG1 is expressed in the petal, an mVenus:PhABCG1 translational fusion was constructed under control of a 2 kbp PhABCG1 promoter (ABCG1:mVenus-ABCG1) for localization in protoplasts originating from petals of the non-fragrant petunia line M1×V30. Since ODO1 is not expressed in petals of M1×V30 (data not shown) and because ODO1 activates the PhABCG1 promoter (Fig. 5), it was decided to co-transform an N-terminal ODO1 translational fusion to GFP under control of the 1.9 kbp ODO1 promoter of cultivar Mitchell (ODO1:GFP–ODO1), which was shown to be activated by EOBII in non-fragrant flowers (Van Moerkercke et al., 2011). By doing so, this allowed identification of the cell layer(s) in which PhABCG1 and/or ODO1 are expressed.
Interestingly, it was found that ODO1 was expressed from its own promoter in protoplasts originating from both epidermis and mesophyll cells, resulting in a nuclear GFP signal in both types of protoplasts (Fig. 8A, G). Accordingly, the promoter of PhABCG1 was active in both types of protoplasts, with plasma membrane localization of the mVenus–ABCG1 fusion protein (Fig. 8A, G). Co-transformation with a 35S-driven mTurquoise-tagged plasma membrane control marker (shown in red; Fig. 8B, H) shows co-localization with mVenus–PhABCG1 at the plasma membrane (Fig. 8D, J). Although, the mVenus and GFP signals were simultaneously detected, the respective fusion proteins could be distinguished based on their different localization, as is illustrated in Figs 6 and 7. In addition, the two signals could be separated using spectral unmixing (data not shown). Autofluorescence of anthocyanins in the vacuole identifies protoplasts originating from epidermal cells (Fig. 8I), which can also be seen in the bright field images (Fig. 8K).
To extend these results, the activity of a transcriptional dsRed fusion construct of the IGS promoter (IGS:dsRed), which like ODO1 is activated by EOBII (Spitzer-Rimon et al., 2010), was tested. The IGS promoter was active in protoplasts originating from mesophyll cells (left in Fig. 9C, I), which do not contain anthocyanins and as a consequence do not autofluoresce upon excitation (Fig. 9D, J). Excitation of dsRed also excites the anthocyanins in the vacuoles of epidermal protoplasts (right in Fig. 9C, I). These two signals could not be detected separately in the confocal set-up used here. However, excitation of anthocyanins did not excite dsRed (Fig. 9D, J). Therefore, a cytosolic dsRed signal can be observed in transformed (Fig. 9E, F), but not in non-transformed (Fig. 9K, L), epidermal protoplasts, showing activation of the IGS promoter in protoplasts originating from epidermal cell layers as well. Together, the results presented in Figs 8 and 9 show that a TF, a biosynthetic gene, and a transporter are active in the different cell types of the petunia petal.
Discussion
Floral volatiles are released from the epidermal cells of the petal limb. Because of their metabolic and ecological relationship with anthocyanins, which accumulate in the petal epidermis, floral volatile production is predicted to occur in the petal epidermis. Indeed, enzymes catalysing the final steps in volatile phenylpropanoid/benzenoid production have been localized predominantly to the petal epidermis in some species such as rose and snapdragon (Kolosova et al., 2001; Scalliet et al., 2006; Bergougnoux et al., 2007; Long et al., 2009), although, to some extent, localization to subepidermal or mesophyll cell layers has been reported for some of them as well (Rohrbeck et al., 2006; Scalliet et al., 2006). Whether this is true for other species and whether the entire pathway is active in these cells, however, has remained unknown. In contrast to this predicted cell specificity, the petunia EPSPS promoter, which is activated by the transcriptional regulator ODO1 (Verdonk et al., 2005), is active in both epidermal and mesophyll cell layers of mature petunia petal limbs (Benfey et al., 1990) and is probably associated with scent production, as it is not involved in anthocyanin or flavonol biosynthesis at this stage of flower development (Koes et al., 1990; Holton et al., 1993; Verdonk et al., 2005). This observation suggests that early steps in the volatile phenylpropanoid/benzenoid production might occur in both cell layers of the petal, and raises questions about the cell-specific activity of other scent-related genes in petunia petals. To test this and to extend the observations made by Benfey et al. (1990), protoplasts derived from M1×V30 petunia petals were used to investigate the activity of the EOBII target genes ODO1 and IGS, as well as the newly identified ODO1 target gene PhABCG1. For a 2 kbp fragment of the FDRa promoter, these protoplasts were recently shown to retain cell-specific activity (Faraco et al., 2011), corresponding to the epidermal localization of the DFRa mRNA (Quattrocchio et al., 2006) and the activity of one of its activators, AN2 (Quattrocchio et al., 1998). This shows that the 2 kbp DFRa promoter region is sufficient for correct expression and that the protoplast system can be a reliable alternative for transcript localization studies such as in situ mRNA hybridizations (Faraco et al., 2011). Previously, the ODO1 promoter was characterized and it was shown that all critical elements for tissue-specific, developmental, and circadian expression are contained within the 1.9 kbp region upstream of the ATG (Van Moerkercke et al., 2011).
Here, it was found that the 1.9 kbp ODO1 promoter is active in both cell layers of the petal (Fig. 8), explaining the observed activity of EPSPS in these cell layers (Benfey et al., 1990) and implying that the activator(s) of ODO1 are active in both cell layers. EOBII, as an activator of ODO1, is thus potentially active in both cell layers as well. However, since EOBII is probably not the only activator of ODO1 (Van Moerkercke et al., 2011), cell specificity cannot be concluded from this result alone. Therefore, the promoter of IGS (Fig. 9), which is also activated by EOBII (Spitzer-Rimon et al., 2010), was tested. IGS is of particular interest since it catalyses a final step in volatile compound biosynthesis in petunia and is thus expected to act in epidermal cells, where volatile release occurs. In addition, transcripts of an isoeugenol-methylating gene, IEMT, were localized predominantly to the epidermis cells in Clarkia breweri (Dudareva and Pichersky, 2000). It was shown that, in addition to ODO1, the promoter of IGS is also activated in both cell layers (Fig. 9). This raises the question of which parts of the pathway are active in which cell layer. The contribution of mesophyll cells to floral volatile phenylpropanoid/benzenoid production seems plausible since petal mesophyll cells have been reported to contain large amounts of sugar, which has been associated with scent production (Bergougnoux et al., 2007). From the present and previous data, it appears that both early and late steps of the pathway occur in both cell layers. Importantly, these experiments suggest that trafficking of (volatile) compounds between these cell layers occurs and that, as a consequence, transport proteins such as ABC transporters might be involved.
With respect to this, a new ODORANT1 target gene was identified that is co-regulated with genes of the floral phenylpropanoid/benzenoid pathway in petunia and that encodes a putative ABC transporter (Fig. 3). Transporters are often co-expressed with enzymes that metabolize the products that need transport. As a consequence, it is likely that they are under control of the same regulators. For instance, the Zea mays multidrug resistance protein 3 (ZmMRP3), encoding an ABC transporter, was shown to transport anthocyanins into the vacuole (Goodman et al., 2004). Importantly, ZmMPR3 is co-expressed with genes of the anthocyanin pathway, and regulators of anthocyanin biosynthesis control ZmMRP3 expression (Goodman et al., 2004). Likewise, the Arabidopsis ABCG26/WBC27, required for exine formation during pollen development, is co-expressed with genes encoding enzymes involved in sporopollenin precursor synthesis (Choi et al., 2010), and ZmWBC11a, involved in cuticular wax secretion, is regulated by the same regulator that activates genes of lipid metabolism (Javelle et al., 2010). PhABCG1 transcripts were exclusively detected in petal tissue (Fig. 1B) and accumulated from anthesis onwards, when floral volatile production starts (Fig. 1C). This excludes a role for PhABCG1 in pigment transport/biosynthesis, since pigments are produced early in flower development (Martin and Gerats, 1993), and is in agreement with the developmental regulation of genes of the volatile benzenoid/phenylpropanoid pathway (Verdonk et al., 2003). Like the emission of floral volatiles, transcription of volatile benzenoid/phenylpropanoid genes in petunia oscillates during the night and day (Colquhoun et al., 2010b). Previously, microarray analysis (Verdonk et al., 2003) revealed rhythmic transcript levels for genes encoding ODO1, 3-ketoacyl-CoA thiolase 1 (KAT1), benzoyl-CoA:benzylaclohol/phenylethylalcohol benzoyl transferase (BPBT), benzoic acid/salicylic acid methyltransferase (BSMT), phenyalanine ammonia lyase (PAL) 1/2, chorismate mutase (CM), and IGS, all of which have later been shown to be involved in floral fragrance biosynthesis (Negre et al., 2003; Boatright et al., 2004; Verdonk et al., 2005; Koeduka et al., 2006; Dexter et al., 2007; Van Moerkercke et al., 2009; Colquhoun et al., 2010a). Importantly, PhABCG1 transcript accumulation oscillates in phase with that of ODO1 (Fig. 1A), which also cycles in phase with mRNA levels of acetyl-CoA:coniferyl alcohol acetyltransferase (CFAT) (Dexter et al., 2007), PAL1 (Colquhoun et al., 2010b), and EPSPS (Verdonk et al., 2003).
As for EPSPS (Verdonk et al., 2005), transcript accumulation and promoter activity of PhABCG1 correlated with ODO1 expression (Figs 2, 4), and transient expression of ODO1 activated the PhABCG1 promoter in leaves and petals (Fig. 5). As a consequence, it was found that PhABCG1, like EPSPS, is activated in both mesophyll and epidermis protoplasts (Fig. 8). In contrast, promoters that are not activated by ODO1, such as the DFRa promoter (Verdonk et al., 2005), are not activated in mesophyll protoplasts (Faraco et al., 2011). A detailed analysis of the PhABCG1 promoter will be needed to identify the ODO1-binding site, as well as to identify additional activators of PhABCG1. The PhABCG1 promoter contains two putative MYB-binding sites (TAACTATCT) within a 39 bp perfect repeat. This element differs in one nucleotide from the MYB.Ph3-binding site (TAACTAACT) found in promoters of flavonoid genes (Solano et al., 1995). However, when deleting this 39 bp repeat in the reporter construct, ODO1 could still activate it (unpublished results), indicating that this putative MYB-binding site is not the binding site for ODO1.
With a growing number of genes identified and recent progress in metabolic engineering, it is becoming increasingly important to identify the inter- and intracellular organization of metabolic pathways. Where and when a transgene is expressed will be important to access control over the metabolic pathway. It has been shown here that the promoter of one master regulator and one of its target genes are active in both cell layers of the petal. Could this mean that the entire pathway is active in both cells? The finding that IGS is active in mesophyll cells is surprising, since it encodes an enzyme that catalyses a final step in the production of the volatile compound isoeugenol. These results suggest (i) that the entire volatile benzenoid/phenylpropanoid pathway is active in all cell layers; (ii) trafficking of metabolites between cell layers of the petal; and (iii) the involvement of transport proteins such as ABC transporters. Transport across the plasma membrane of some intermediates of isoeugenol biosynthesis, which are also intermediates in lignin production, has been shown to be ATP dependent (Kaneda et al., 2011). Also benzoic acid export in Schizosaccharomyces pombe and Arabidopsis roots can be facilitated by plant ABC proteins (Yang and Murphy, 2009; Kubes et al., 2011). How volatile compounds are emitted from petals after biosynthesis remains unknown and could involve active transport, diffusion, or both (Jetter, 2006). Several reports suggest the involvement of transport proteins in fragrance emission, although conflicting observations exist. A decrease in endogenous concentrations and an increase in emission of volatile compounds from flowers of petunia were reported for increasing temperatures. At lower temperatures, differences in the ratio of the emitted and endogenous concentration between the compounds become more apparent, meaning that at low temperatures, the emission of large compounds is more hampered than that of small compounds. This suggests a physical component of volatile emission and opens up the possibility of passive diffusion of these compounds (Sagae et al., 2008). In addition, the emission of compounds follows the endogenous concentration during flower development (Goodwin et al., 2003). This in contrast to the finding that emission of a compound can be affected without affecting internal levels, or vice versa. For example, overexpression of the R2R3 MYB PAP1 in petunia petals does not alter internal levels of benzaldehyde, whereas emission of this compound increases; moreover, internal levels of phenylacetaldehyde increase without being emitted (Ben Zvi et al., 2008). PhABCG1 clusters with uncharacterized ABC proteins, rendering it impossible to predict its function based on the sequence. The closest homologue is AtWBC11, which is involved in cuticle formation. The role of PhABCG1 in floral scent production is currently being investigated, taking a genetic approach. The experiments presented here assess the cell specificity of fragrance production in petunia petals and suggest fragrance production in both epidermal and mesophyll cells of the petal based on the promoter activity of (i) a transcriptional regulator; (ii) a biosynthetic enzyme catalysing an end-product; and (iii) a newly identified transporter. Evidence on the localization of other genes and proteins of the pathway, in petunia and other species, will be needed to construct a generalized and integrated model for fragrance biosynthesis in petals. A challenging issue to address next is the relative contribution of each cell layer. Quantitative strategies, such as in situ hybridization or laser capture microdissection coupled to quantitative RT-PCR, could be used to evaluate this.
Acknowledgments
We wish to thank Alexander Vainstein for sharing the IGS:dsRed construct, Anna Pietraszewska for the mTurquoise membrane marker, Marianna Faraco for help with petal protoplast isolation, Francesca Quattrocchio for the M1×V30 plants and 35S:AN2 construct, and Christa Testerink for sharing the GW destination vector. Confocal laser scanning microscopy was performed at the Centre for Advanced Microscopy (CAM) of the University of Amsterdam and we wish to acknowledge them for the use of their equipment. Ludek Tikovsky, Harold Lemereis, and Thijs Hendrix are acknowledged for taking care of the petunia plants. We thank two reviewers for helpful comments and suggestions.
References
- Ben Zvi MM, Florence NZ, Masci T, Ovadis M, Shklarman E, Ben-Meir H, Tzfira T, Dudareva N, Vainstein A. Interlinking showy traits: co-engineering of scent and colour biosynthesis in flowers. Plant Biotechnology Journal. 2008;6:403–415. doi: 10.1111/j.1467-7652.2008.00329.x. [DOI] [PubMed] [Google Scholar]
- Benfey PN, Takatsuji H, Ren L, Shah DM, Chua NH. Sequence requirements of the 5-enolpyruvylshikimate-3-phosphate synthase 5′-upstream region for tissue-specific expression in flowers and seedlings. The Plant Cell. 1990;2:849–856. doi: 10.1105/tpc.2.9.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergougnoux V, Caissard JC, Jullien F, Magnard JL, Scalliet G, Cock JM, Hugueney P, Baudino S. Both the adaxial and abaxial epidermal layers of the rose petal emit volatile scent compounds. Planta. 2007;226:853–866. doi: 10.1007/s00425-007-0531-1. [DOI] [PubMed] [Google Scholar]
- Bird D, Beisson F, Brigham A, Shin J, Greer S, Jetter R, Kunst L, Wu X, Yephremov A, Samuels L. Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. The Plant Journal. 2007;52:485–498. doi: 10.1111/j.1365-313X.2007.03252.x. [DOI] [PubMed] [Google Scholar]
- Boatright J, Negre F, Chen X, Kish CM, Wood B, Peel G, Orlova I, Gang D, Rhodes D, Dudareva N. Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiology. 2004;135:1993–2011. doi: 10.1104/pp.104.045468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Talbot C, Boutte Y, Catrice O, Read ND, Satiat-Jeunemaitre B. FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. Journal of Microscopy. 2004;214:159–173. doi: 10.1111/j.0022-2720.2004.01348.x. [DOI] [PubMed] [Google Scholar]
- Choi H, Jin JY, Choi S, Hwang JU, Kim YY, Suh MC, Lee Y. An ABCG/WBC-type ABC transporter is essential for transport of sporopollenin precursors for exine formation in developing pollen. The Plant Journal. 2010;65:181–193. doi: 10.1111/j.1365-313X.2010.04412.x. [DOI] [PubMed] [Google Scholar]
- Colquhoun TA, Schimmel BC, Kim JY, Reinhardt D, Cline K, Clark DG. A petunia chorismate mutase specialized for the production of floral volatiles. The Plant Journal. 2010a;61:145–155. doi: 10.1111/j.1365-313X.2009.04042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun TA, Verdonk JC, Schimmel BC, Tieman DM, Underwood BA, Clark DG. Petunia floral volatile benzenoid/phenylpropanoid genes are regulated in a similar manner. Phytochemistry. 2010b;71:158–167. doi: 10.1016/j.phytochem.2009.09.036. [DOI] [PubMed] [Google Scholar]
- Dexter R, Qualley A, Kish CM, Ma CJ, Koeduka T, Nagegowda DA, Dudareva N, Pichersky E, Clark D. Characterization of a petunia acetyltransferase involved in the biosynthesis of the floral volatile isoeugenol. The Plant Journal. 2007;49:265–275. doi: 10.1111/j.1365-313X.2006.02954.x. [DOI] [PubMed] [Google Scholar]
- Dudareva N, Pichersky E. Biochemical and molecular genetic aspects of floral scents. Plant Physiology. 2000;122:627–633. doi: 10.1104/pp.122.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Pichersky E. Metabolic engineering of plant volatiles. Current Opinion in Biotechnology. 2008;19:181–189. doi: 10.1016/j.copbio.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Faraco M, Di Sansebastiano GP, Spelt K, Koes RE, Quattrocchio FM. One protoplast is not the other! Plant Physiology. 2011;156:474–478. doi: 10.1104/pp.111.173708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhart J, van Weeren L, Hink MA, Vischer NO, Jalink K, Gadella TW., Jr Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nature Methods. 2010;7:137–139. doi: 10.1038/nmeth.1415. [DOI] [PubMed] [Google Scholar]
- Goodman CD, Casati P, Walbot V. A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. The Plant Cell. 2004;16:1812–1826. doi: 10.1105/tpc.022574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin SM, Kolosova N, Kish CM, Wood KV, Dudareva N, Jenks MA. Cuticle characteristics and volatile emissions of petals in Antirrhinum majus. Physiologia Plantarum. 2003;117:435–443. doi: 10.1034/j.1399-3054.2003.00047.x. [DOI] [PubMed] [Google Scholar]
- Higgins CF, Hiles ID, Salmond GP, et al. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature. 1986;323:448–450. doi: 10.1038/323448a0. [DOI] [PubMed] [Google Scholar]
- Holton TA, Brugliera F, Tanaka Y. Cloning and expression of flavonol synthase from Petunia hybrida. The Plant Journal. 1993;4:1003–1010. doi: 10.1046/j.1365-313x.1993.04061003.x. [DOI] [PubMed] [Google Scholar]
- Ivanchenko M, Vejlupkova Z, Quatrano RS, Fowler JE. Maize ROP7 GTPase contains a unique, CaaX box-independent plasma membrane targeting signal. The Plant Journal. 2000;24:79–90. doi: 10.1046/j.1365-313x.2000.00855.x. [DOI] [PubMed] [Google Scholar]
- Jasinski M, Stukkens Y, Degand H, Purnelle B, Marchand-Brynaert J, Boutry M. A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. The Plant Cell. 2001;13:1095–1107. [PMC free article] [PubMed] [Google Scholar]
- Javelle M, Vernoud V, Depege-Fargeix N, Arnould C, Oursel D, Domergue F, Sarda X, Rogowsky PM. Overexpression of the epidermis-specific homeodomain-leucine zipper IV transcription factor Outer Cell Layer1 in maize identifies target genes involved in lipid metabolism and cuticle biosynthesis. Plant Physiology. 2010;154:273–286. doi: 10.1104/pp.109.150540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetter R. Examination of the processes involved in the emission of scent volatiles from flowers. In: Dudareva N, Pichersky E, editors. Biology of floral scent. Boca Raton, FL: Taylor and Francis Group; 2006. pp. 125–144. [Google Scholar]
- Kaminaga Y, Schnepp J, Peel G, et al. Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation. Journal of Biological Chemistry. 2006;281:23357–23366. doi: 10.1074/jbc.M602708200. [DOI] [PubMed] [Google Scholar]
- Kaneda M, Schuetz M, Lin BS, Chanis C, Hamberger B, Western TL, Ehlting J, Samuels AL. ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. Journal of Experimental Botany. 2011;62:2063–2077. doi: 10.1093/jxb/erq416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeduka T, Fridman E, Gang DR, et al. Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proceedings of the National Academy of Sciences, USA. 2006;103:10128–10133. doi: 10.1073/pnas.0603732103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes RE, Van Blokland R, Quattrocchio F, Van Tunen AJ, Mol J. Chalcone synthase promoters in petunia are active in pigmented and unpigmented cell types. The Plant Cell. 1990;2:379–392. doi: 10.1105/tpc.2.5.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends in Plant Science. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Kolosova N, Sherman D, Karlson D, Dudareva N. Cellular and subcellular localization of S-adenosyl-methionine:benzoic acid carboxyl methyltransferase, the enzyme responsible for biosynthesis of the volatile ester methylbenzoate in snapdragon flowers. Plant Physiology. 2001;126:956–964. doi: 10.1104/pp.126.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubes M, Yang H, Richter GL, et al. 2011. The Arabidopsis concentration-dependent influx/efflux transporter ABCB4 regulates cellular auxin levels in the root epidermis. The Plant Journal. doi: 10.1111/j.1365-313X.2011.04818.x. (in press) [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lee MW, Yang Y. Transient expression assay by agroinfiltration of leaves. Methods in Molecular Biology. 2006;323:225–229. doi: 10.1385/1-59745-003-0:225. [DOI] [PubMed] [Google Scholar]
- Long MC, Nagegowda DA, Kaminaga Y, Ho KK, Kish CM, Schnepp J, Sherman D, Weiner H, Rhodes D, Dudareva N. Involvement of snapdragon benzaldehyde dehydrogenase in benzoic acid biosynthesis. The Plant Journal. 2009;59:256–265. doi: 10.1111/j.1365-313X.2009.03864.x. [DOI] [PubMed] [Google Scholar]
- Maeda H, Shasany AK, Schnepp J, Orlova I, Taguchi G, Cooper BR, Rhodes D, Pichersky E, Dudareva N. RNAi suppression of Arogenate Dehydratase1 reveals that phenylalanine is synthesized predominantly via the arogenate pathway in petunia petals. The Plant Cell. 2010;22:832–849. doi: 10.1105/tpc.109.073247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Gerats T. Control of pigment biosynthesis genes during petal development. The Plant Cell. 1993;5:1253–1264. doi: 10.1105/tpc.5.10.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao YC, Liu CJ. ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proceedings of the National Academy of Sciences, USA. 2010;107:22728–22733. doi: 10.1073/pnas.1007747108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nature Biotechnology. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Negre F, Kish CM, Boatright J, Underwood B, Shibuya K, Wagner C, Clark DG, Dudareva N. Regulation of methylbenzoate emission after pollination in snapdragon and petunia flowers. The Plant Cell. 2003;15:2992–3006. doi: 10.1105/tpc.016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikashvili D, Savaldi-Goldstein S, Mandel T, Yifhar T, Franke RB, Hofer R, Schreiber L, Chory J, Aharoni A. The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiology. 2007;145:1345–1360. doi: 10.1104/pp.107.105676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikashvili D, Shi JX, Schreiber L, Aharoni A. The Arabidopsis ABCG13 transporter is required for flower cuticle secretion and patterning of the petal epidermis. New Phytologist. 2011;190:113–124. doi: 10.1111/j.1469-8137.2010.03608.x. [DOI] [PubMed] [Google Scholar]
- Pighin JA, Zheng H, Balakshin LJ, Goodman IP, Western TL, Jetter R, Kunst L, Samuels AL. Plant cuticular lipid export requires an ABC transporter. Science. 2004;306:702–704. doi: 10.1126/science.1102331. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Verweij W, Kroon A, Spelt C, Mol J, Koes R. PH4 of Petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix–loop–helix transcription factors of the anthocyanin pathway. The Plant Cell. 2006;18:1274–1291. doi: 10.1105/tpc.105.034041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, Leppen H, Mol J, Koes RE. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. The Plant Cell. 1993;5:1497–1512. doi: 10.1105/tpc.5.11.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, van der Woude K, Mol JN, Koes R. Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. The Plant Journal. 1998;13:475–488. doi: 10.1046/j.1365-313x.1998.00046.x. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R. Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. The Plant Cell. 1999;11:1433–1444. doi: 10.1105/tpc.11.8.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrbeck D, Buss D, Effmert U, Piechulla B. Localization of methyl benzoate synthesis and emission in Stephanotis floribunda and Nicotiana suaveolens flowers. Plant Biology (Stuttgart, Germany) 2006;8:615–626. doi: 10.1055/s-2006-924076. [DOI] [PubMed] [Google Scholar]
- Sagae M, Oyama-Okubo N, Ando T, Marchesi E, Nakayama M. Effect of temperature on the floral scent emission and endogenous volatile profile of Petunia axillaris. Bioscience, Biotechnology, and Biochemistry. 2008;72:110–115. doi: 10.1271/bbb.70490. [DOI] [PubMed] [Google Scholar]
- Scalliet G, Lionnet C, Le Bechec M, et al. Role of petal-specific orcinol O-methyltransferases in the evolution of rose scent. Plant Physiology. 2006;140:18–29. doi: 10.1104/pp.105.070961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurink RC, Haring MA, Clark DG. Regulation of volatile benzenoid biosynthesis in petunia flowers. Trends in Plant Science. 2006;11:20–25. doi: 10.1016/j.tplants.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Research. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Nieto C, Avila J, Canas L, Diaz I, Paz-Ares J. Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB.Ph3) from Petunia hybrida. EMBO Journal. 1995;14:1773–1784. doi: 10.1002/j.1460-2075.1995.tb07166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer-Rimon B, Marhevka E, Barkai O, Marton I, Edelbaum O, Masci T, Prathapani NK, Shklarman E, Ovadis M, Vainstein A. EOBII, a gene encoding a flower-specific regulator of phenylpropanoid volatiles’ biosynthesis in petunia. The Plant Cell. 2010;22:1961–1976. doi: 10.1105/tpc.109.067280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanengelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ. Pbinplus—an improved plant transformation vector based on Pbin19. Transgenic Research. 1995;4:288–290. doi: 10.1007/BF01969123. [DOI] [PubMed] [Google Scholar]
- Van Moerkercke A, Haring MA, Schuurink RC. The transcription factor EMISSION OF BENZENOIDS II activates the MYB ODORANT1 promoter at a MYB binding site specific for fragrant petunias. The Plant Journal. 2011;67:917–928. doi: 10.1111/j.1365-313X.2011.04644.x. [DOI] [PubMed] [Google Scholar]
- Van Moerkercke A, Schauvinhold I, Pichersky E, Haring MA, Schuurink RC. A plant thiolase involved in benzoic acid biosynthesis and volatile benzenoid production. The Plant Journal. 2009;60:292–302. doi: 10.1111/j.1365-313X.2009.03953.x. [DOI] [PubMed] [Google Scholar]
- Verdonk JC. PhD thesis. University of Amsterdam; 2006. Floral scent production by Petunia hybrida. [Google Scholar]
- Verdonk JC, Haring MA, van Tunen AJ, Schuurink RC. ODORANT1 regulates fragrance biosynthesis in petunia flowers. The Plant Cell. 2005;17:1612–1624. doi: 10.1105/tpc.104.028837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdonk JC, Ric de Vos CH, Verhoeven HA, Haring MA, van Tunen AJ, Schuurink RC. Regulation of floral scent production in petunia revealed by targeted metabolomics. Phytochemistry. 2003;62:997–1008. doi: 10.1016/s0031-9422(02)00707-0. [DOI] [PubMed] [Google Scholar]
- Verrier PJ, Bird D, Burla B, et al. Plant ABC proteins—a unified nomenclature and updated inventory. Trends in Plant Science. 2008;13:151–159. doi: 10.1016/j.tplants.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Verweij W, Spelt C, Di Sansebastiano GP, Vermeer J, Reale L, Ferranti F, Koes R, Quattrocchio F. An H+ P-ATPase on the tonoplast determines vacuolar pH and flower colour. Nature Cell Biology. 2008;10:1456–1462. doi: 10.1038/ncb1805. [DOI] [PubMed] [Google Scholar]
- Vogt T. Phenylpropanoid biosynthesis. Molecular Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO Journal. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Murphy AS. Functional expression and characterization of Arabidopsis ABCB, AUX 1 and PIN auxin transporters in Schizosaccharomyces pombe. The Plant Journal. 2009;59:179–191. doi: 10.1111/j.1365-313X.2009.03856.x. [DOI] [PubMed] [Google Scholar]