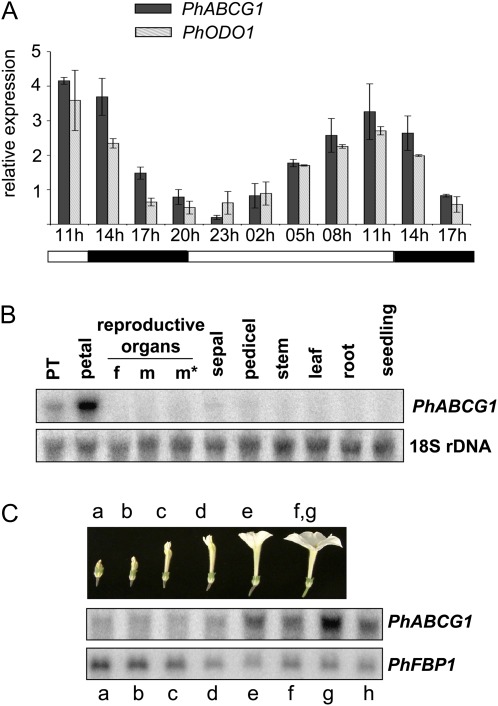
Fig. 1.
Characterization of PhABCG1 expression. RNA gel blots were hybridized with the indicated probes. Re-hybridization with a FLORAL BINDING PROTEIN 1 (FBP1) or an 18S rDNA probe was used to show loading of the gel or to enable normalization. (A) Rhythmic expression of PhABCG1 in Mitchell flowers, in phase with ODO1 expression and rhythmic volatile emission (not shown; Verdonk et al., 2003). Mean values are shown (n=2). Error bars indicate maximum and minimum values. Black and grey bars indicate PhABCG1 and ODO1 transcript levels, respectively. Petal tissue was harvested at the time points indicated below the graph (3 h intervals). Black and white bars below the graph indicate dark and light conditions, respectively. (B) Tissue-specific accumulation in petunia Mitchell. The tissues used are indicated (f, style and stigma; m, non-dehisced anthers; m*, dehisced anthers; PT, petal tube; petal refers to the petal limb). (C) Developmentally regulated transcript accumulation in Mitchell petals. Developmental stages of the petals: a–e, as indicated; f and g indicate open flowers before and after anthesis, respectively; h indicates a senescing flower.