Abstract
Mismatch repair (MMR) is an evolutionarily conserved DNA repair system, which corrects mismatched bases arising during DNA replication. MutS recognizes and binds base pair mismatches, while the MutL protein interacts with MutS–mismatch complex and triggers MutH endonuclease activity at a distal-strand discrimination site on the DNA. The mechanism of communication between these two distal sites on the DNA is not known. We used functional fluorescent MMR proteins, MutS and MutL, in order to investigate the formation of the fluorescent MMR protein complexes on mismatches in real-time in growing Escherichia coli cells. We found that MutS and MutL proteins co-localize on unrepaired mismatches to form fluorescent foci. MutL foci were, on average, 2.7 times more intense than the MutS foci co-localized on individual mismatches. A steric block on the DNA provided by the MutHE56A mutant protein, which binds to but does not cut the DNA at the strand discrimination site, decreased MutL foci fluorescence 3-fold. This indicates that MutL accumulates from the mismatch site toward strand discrimination site along the DNA. Our results corroborate the hypothesis postulating that MutL accumulation assures the coordination of the MMR activities between the mismatch and the strand discrimination site.
INTRODUCTION
Since the first experiment that showed a direct evidence for the existence of a strand-directed mismatch repair (MMR) in bacteria in 1976 (1), several thousand papers describing MMR in vivo and in vitro have been published (2–5). MMR is a major guardian of genomic stability that improves the fidelity of DNA replication (6), prevents genomic rearrangements (7) and acts as genetic barrier between related species (8) by abolishing recombination between non-identical DNA sequences (2,6). Key bacterial MMR proteins, MutS and MutL, are conserved in all kingdoms of life (2,6). The MutS protein, and its eukaryotic homologs, detects and binds diverse mismatches arising during DNA replication. The MutL protein, and its eukaryotic homologs, links a mismatch-bound MutS with the strand discrimination site of helicase/exonuclease activities that remove the mismatched base specifically from the newly synthesized strand.
In Escherichia coli, discrimination between template and newly synthesized strand is based on the methylation status of adenines in the GATC sequences (2,6). In newly synthesized strands, adenines in GATC sequences are transiently unmethylated thus providing a binding site for the MutH endonuclease. MutH is a MutL-binding protein that can cleave the newly synthesized strand at a proximal non-methylated GATC sequence on either side of the mismatch (2,6). DNA cleavage by MutH triggers mismatch repair involving UvrD (helicase II), single-stranded exonucleases, DNA polymerase III holoenzyme, single-strand DNA-binding protein and DNA ligase (2,6). In E. coli, GATC cleavage can occur even at kilobase or larger distances from the mismatch (6,9).
Despite the decades of research, a key mechanistic aspect of mismatch repair remained mysterious, i.e. what is the nature of molecular communication between the site of mismatch and the site of strand discrimination? Several conceptually exclusive models, which can be grouped into three categories, have been proposed to explain interaction between these two DNA sites. All models are based exclusively on in vitro biochemical studies of MMR proteins and there are yet no in vivo experiments that confirm their biological relevance. The first model postulates that DNA looping brings the stationary mismatch–MutS–MutL complex and the hemi-methylated GATC sequence in proximity (10–12). The second model proposes that MutS–MutL complex, or MutS alone, travel along the DNA helix from the mismatch towards the proximal strand discrimination site (13,14). Finally, the third model suggests that mismatch recognition by MutS triggers polymerization of MutL along DNA between the mismatch and the strand discrimination signal (15). This model was promulgated by studies showing that MutS binding to the mismatch protects ∼10 bp on either side of the mismatch against DNase I digestion, while the addition of MutL extends the footprint dramatically (11,16). Also the study of the effect of the DNA chain length on the formation of the ternary complex mismatch–MutS–MutL led to the proposal that the ternary complex formation could involve the polymerization of MutL along the helix (17). A corollary of this model is the assumption that more MutL than MutS proteins should be required for the repair of a mismatch. This was further strengthened by genetic evidence showing that the cellular level of MutL, and not of MutS, becomes limiting when MMR is saturated in the presence of numerous mismatches (18–22). Because both MutS and MutL are present in equimolar concentrations in the cell, MutL protein limitation cannot simply be due to a lower amount of MutL in the cell (23).
In this study, we looked whether more MutL than MutS proteins are present on the mismatches in vivo. We investigated individual growing E. coli cells carrying functional MutS and MutL proteins fused to fluorescent proteins and followed the formation of the fluorescent MMR protein complexes on individual mismatches in real-time by fluorescent microscopy. Our previously published data show that the MutL forms fluorescent foci on unrepairable mismatches and that MutL foci formation is dependent on MutS (24). However, the expected co-localization of MutS and MutL was not explored and the relative amount of MutS and MutL on these mismatches was not quantified. Here, we show that MutL and MutS fluorescent foci co-localize and that the fluorescence of MutL foci is always more intense than the fluorescence of MutS foci. Furthermore, the presence of a roadblock at the GATC sequences reduces the amount of MutL on the mismatch. This indicates that the in vivo accumulation of the MutL protein around mismatch is spanning the mismatch site and the site of strand discrimination. In conclusion, our results corroborate the hypothesis postulating that MutL polymerization could account for the communication between two distant DNA sites involved in MMR.
MATERIALS AND METHODS
Escherichia coli strains expressing fluorescently tagged MutL and MutS
To assess the co-localization of MutS and MutL proteins in single E. coli cells, we constructed translational fusions of Yellow Fluorescent Protein (YFP) and Cyan Fluorescent Protein (CFP) to the N-termini of MutS and MutL proteins. This was done by modifying the plasmids peGFP-MutSwt and peGFP-MutLwt (24) in several steps. First, we replaced eGFP in the peGFP-MutLwt and peGFP-MutSwt by YFP and CFP. We amplified by PCR the fragment encoding YFP from the template pUC18-yfp (25) and the fragment encoding CFP from the template pUC18-cfp (26). We used primers 5′CCCAGATCTGGGTACCGACGACGACGACAAGATGGCTAGCAAAGGAGAACTTTTCAC-3 and 5′ATGAATTCGCCAGATCCTGATCCTGATCCAGATCCTGAGCCGCTTGGTAGAGCTCATCCATGCCATGTG-3 for yfp amplification. For cfp amplification, we used primers 5′CCCAGATCTGGGTACCGACGACGACGACAAGATGGCTAGCAAAGGAGAAGAACTTTTCAC-3′ and 5′TGAATTCGCCAGATCCTGATCCAGATCCTGAGCCGCTGATATCATCTTTGTAGAGCTCATCCATGCCATGTG-3′. These primer pairs include a BglII restriction site at the N-terminal and an EcoRI (for yfp) or an EcoRV restriction site (for cfp) at the C-terminal. We digested the PCR-amplified fragments by BglII and by EcoRI or EcoRV and cloned them in the peGFP-MutSwt or peGFP-MutLwt. This resulted in the plasmids pYFP-MutS, pCFP-MutS, pYFP-MutL and pCFP-MutL expressing YFP-MutS, CFP-MutS, YFP-MutL and CFP-MutL, respectively. In the second step, we cloned the compatible fluorescent versions of mutL and mutS on the same plasmid, each under control of the individual T7 RNA polymerase promoter. We used the plasmids pCFP-MutS and pCFP-MutL as templates for PCR amplification of the fragments encoding CFP-MutS and CFP-MutL and including T7 RNA polymerase promoter and terminator. We used for PCR amplification primers 5′-GGGCATGCTAATACGACTCACTATAGGG-3′ and 5′-GGGCATGCCAAAAAACCCTCAAGACCCG-3′ with a SphI restriction site at the N- and C-termini. We digested PCR products by SphI and cloned the digested fragment in SphI site of the pYFP-MutL or pYFP-MutS. This resulted in the plasmid pYFP-MutL CFP-MutS expressing YFP-MutL and CFP-MutS, and the plasmid pYFP-MutS CFP-MutL expressing YFP-MutS and CFP-MutL. These plasmids were introduced in cells with inactivated chromosomal mutS and mutL genes.
To carry out experiments in which we determine the effect of the roadblock protein MutHE56A on MutL focus fluorescence, we cloned wild-type or partial loss-of-function mutH (mutHE56A) allele on the pBAD24 plasmid under arabinose promoter (27) and expressed yfp tagged mutL from lactose promoter on the chromosome.
To construct pBAD24 plasmids expressing MutHwt or MutHE56A, we amplified by PCR the fragment encoding MutHwt using pTX417 as a template, and the fragment encoding MutHE56A using as a template pWY1016 (28,29). We used for PCR amplification primers 5′-GCGAATTCATGTCCCAACCTCGCCCACT-3′ and 5′-GGAAGCTTCTACTGGATCAGAAAATGAC-3′ with an EcoRI restriction site at the N-terminal and a HindIII restriction site at the C-terminal. We digested the obtained PCR products by EcoRI and HindIII and cloned them in the pBAD24. This resulted in the plasmids pBAD24MutHwt or pBAD24MutHE56A expressing MutHwt or MutHE56A.
For yfp-mutL integration in the chromosome, we first modified the plasmid pYFP-MutL to include the chloramphenicol resistance cassette (CmR), flanked by FLP recombinase recognition targets, downstream of mutL. We amplified the sequence coding for the CmR (cat) by PCR using as a template the plasmid pKD3 (30). We used for PCR amplification primers 5′-CCGCGGCCGCGTGTAGGCTGGAGCTGCTTC-3′ and 5′-CCGCGGCCGCCATATGAATATCCTCCTTAG-3′ including a NotI restriction site at the N- and C-termini. We digested the resulting PCR product by NotI and cloned it in NotI site of the pYFP-MutL. This resulted in the plasmid pYFP-MutLCmR. In a second step, we used the pYFP-MutLCmR as a template to clone the yfp-mutL-cat under lactose promoter on the chromosome using gene replacement technique (30). We amplified the yfp-mutL-cat by PCR using primers that include homologies to the chromosomal lactose region. The primer 1 (5′-TGTGTGGAATTGTGAGCGGATAACAATTTCACACAGGAAACAGCTATGGCTAGCAAAGGAGAAGAAC-3′) includes at 5′ 45 bp sequence homologous to lactose promoter region (from −1 to −45) and the primer 2 (5′- CACCAGACCAACTGGTAATGGTAGCGACCGGCGCTCAGCTGTTAGCAGCCGGATCTCAGTG-3′) includes at 5′ 40 bp sequence homologous to lacZ C terminal (from 3026 to 3065). The resulting PCR product was introduced by transformation in MG1655 mutL218::Tn10 strain containing the plasmid pKD46 (30). The selected chloramphenicol resistant lactose− clones were named ME120.
Strains, growth conditions and mutation frequency assays
Strains and plasmids are listed in Supplementary Tables S1 and S2. All strains were derived from wild-type E. coli MG1655 strain using P1 transduction and transformation. We verified strain genotypes by testing the UV resistance, capacity to generate mutations conferring resistance to rifampicin or ability to use arabinose in McConkey plates supplemented by arabinose. We grew cells in M9 medium (31) supplemented by 2 mM MgSO4, 0.003% vitamin B1, 0.001% uracile, 0.2% casamino acids, 0.01% glycerol and ampicillin (100 μg/ml) (to select for plasmids expressing the fluorescently labeled derivatives of MMR proteins). We added arabinose (0.02%) to the growth medium to induce the expression of MutHE56A or MutHwt above their basal level of expression from the pBAD24 plasmid. To block DNA replication we used rifampicin at 150 μg/ml. We performed mutagenesis experiments as described previously (24).
Live cell microscopy
Microscopy of live cells was done as described previously (24). Briefly, we grew cells in supplemented M9 medium to early exponential phase (OD600 0.1–0.2) at 37°C. When indicated, cultures were split in two. We treated one culture with rifampicin for 30 min, while the other culture was not treated. We concentrated the aliquots of treated and untreated cells and spread them on supplemented M9 agarose medium, in a cavity slide to obtain a cell monolayer, as described previously (24). We mounted the slide on Metamorph software (Universal Imaging) driven temperature controlled (Life Imaging Services) Zeiss 200M (Zeiss) inverted microscope. Images were recorded at 100-fold (for MutS/MutL co-localization experiments and for MutL foci fluorescence experiments of rifampicin treated or untreated cells) or 63-fold magnification (for roadblock experiments) using CoolSNAP HQ camera (Princton Instruments), in phase contrast and in fluorescence. Co-localization experiments and MutL foci fluorescence experiments of rifampicin treated or untreated cells we done using HBO 103 lamp (Zeiss) regulated to 100% power at wavelengths of 500 nm (YFP) and 420 nm (CFP) during 20 s (for YFP and eGFP) and 6 s (for CFP) of exposure time. Roadblock experiments were done with HXP 120 lamp (Zeiss) regulated to 100% power at wavelength of 500 nm (YFP) during 6 s of exposure time. We acquired, merged and analyzed images using Metamorph software (Meta Imaging Systems).
Image analysis
Images were analyzed as described previously (24). Briefly, for all foci, we defined the region delimiting each focus, and one control region in the cell, representing the cytoplasmic fluorescence, using Metamorph region tool. For the co-localization analysis, we defined two regions (focus and control) first on the fluorescent image taken with the yellow filter. That was the image that we always recorded first. We then transferred both regions to the same position on the fluorescent image taken with the blue filter and measured the fluorescence of the control and focus regions for each cell, and for each image. We also recorded the fluorescence background of the agarose for each image. We calculated focus fluorescence by subtracting from the maximal pixel intensity of focus region the average fluorescence of control region. The cytoplasmic fluorescence was determined by subtracting from the average fluorescence of the control region the average fluorescence of the background. All presented data are the cumulative from at least three independent experiments, which always showed similar results.
RESULTS
Fluorescently tagged MutS and MutL are functional
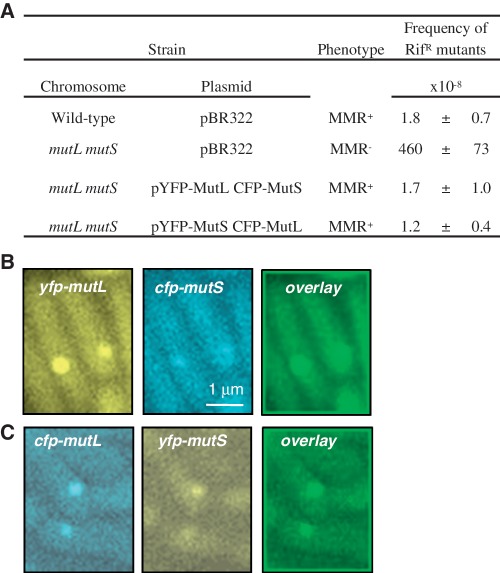
Using GFP-MutL fusion, we and Walker's group showed previously that MutL forms MutS-dependent fluorescent foci on mismatches in growing E. coli and Bacillus subtilis cells (24,32). While the B. subtilis fluorescent MutL was non-functional, the E. coli one that we constructed conserved its function completely. For this study, we constructed YFP- and CFP-tagged MutL and MutS. To determine whether these fluorescent protein fusions are functional, we examined their ability to complement the deletions of mutS and mutL genes (Figure 1A and Supplementary Table S3). The strains deleted for mutL and mutS carrying the plasmid expressing YFP-MutL and CFP-MutS, or carrying the plasmid expressing CFP-MutL and YFP-MutL, exhibited the spontaneous mutagenesis levels of the wild-type strain (Figure 1A). On the other hand, the frequency of mutations of same mutL and mutS strains carrying only the empty vector was 200-fold higher than of the wild-type strain (Figure 1A).
Figure 1.
Functional fluorescent MutL and MutS co-localize on DNA mismatches. (A) Mutation frequency of mutL mutS deletion strain expressing fluorescently labeled MutS and MutL. Fluorescent images (taken with yellow and blue filters) and overlay image of mutH strain expressing (B) YFP-MutL and CFP-MutS or (C) YFP-MutS and CFP-MutL. Mean and standard error of the mean are presented in panel A. RifR stands for rifampicin resistant.
MutL and MutS co-localize on mismatches
To investigate the mismatch repair reaction in vivo, we started by examining the co-localization of MutS and MutL. To do this, we used a mismatch repair deficient mutH strain to get higher number of mismatches in order to have better statistics. The frequency of cells bearing MutL foci in the mutH strain increased from 0.45% in the wild-type strain to 24.9% (24). The observed MutL foci are specific to mismatches because they are not formed without functional MutS protein (24). When we examined MutL foci, we found that they are co-localized with MutS foci. We quantified the frequency of co-localization and found that in 90% of cases MutL foci are co-localized with MutS foci (Figure 1B and C). Since no MutL foci form in the absence of a functional MutS, we conclude that in the remaining 10% of cases MutS foci must be below the threshold of detection (see also below).
Relative stoichiometry of MutL and MutS
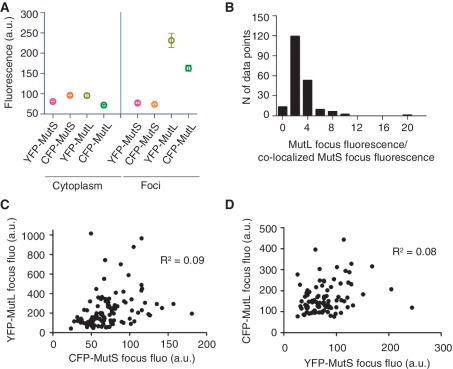
When the number of mismatches increases in the cell, MutL but not MutS becomes limiting (18–22). MutL could become limiting during MMR if the stoichiometry of the mismatch repair reaction is such that more MutL than MutS proteins are needed in the repair of a mismatch. This hypothesis predicts that the MutS and MutL foci co-localized on the mismatches should contain a greater amount of MutL than MutS protein. To compare the relative amount of MutS and MutL proteins per mismatch, we analyzed the fluorescence intensity of the co-localized MutS and MutL foci. First, we identified a MutL focus and the coincident MutS focus, and then we measured their respective fluorescent intensities. We performed these experiments with the combinations of YFP-MutL and CFP-MutS as well as with CFP-MutL and YFP-MutS protein fusions (Figure 1B and C). In both cases, the fluorescence intensity of MutL foci was higher compared to the intensity of MutS foci (Figure 2A). The average ratio of MutL foci fluorescence to co-localized MutS foci fluorescence was 2.7 (Figure 2B). These differences are not due to differences in the cytoplasmic fluorescence that varies very little among cells carrying differently labeled MutS and MutL proteins (Figure 2A).
Figure 2.
The stoichiometry of MutL and MutS on DNA mismatches. (A) The mean cytoplasmic fluorescence and the mean fluorescence of foci for 108 cells with YFP-MutL and CFP-MutS foci from 1B and 95 cells with YFP-MutS and CFP-MutL foci from 1C. (B) The histogram of the ratio of MutL focus fluorescence to co-localized MutS focus fluorescence for 203 pairs of co-localized MutL and MutS foci from A. (C) No correlation between the fluorescence intensity of co-localized foci of YFP-MutL and CFP-MutS (R2 = 0.09) and (D) YFP-MutS and CFP-MutL (R2 = 0.08). Error bars in A indicate standard error of the mean, N in B stands for number, a.u. in A, C and D stands for arbitrary units and fluo in C and D stands for fluorescence.
For the YFP-MutL CFP-MutS pairs, the mean YFP-MutL and CFP-MutS foci fluorescence (±SEM) was 230 (±17.6) and 73 (±2.7), respectively. For the YFP-MutS CFP-MutL pairs, the mean YFP-MutS and CFP-MutL foci fluorescence was 76 (±3.8) and 162 (±7.4), respectively. Observed differences between fluorescence intensity of MutL and MutS foci are statistically significant (P < 0.0001, Wilcoxon test).
To examine the possible stoichiometric relationships of MutS and MutL co-assembled on mismatches, we examined the correlation between the intensity of fluorescence of the co-localized MutL and MutS in single foci. We detected no correlation between the MutL and MutS fluorescent intensities. For the YFP-MutL CFP-MutS pair, the R2 was 0.09 (Figure 2C) and for the YFP-MutS CFP-MutL pair R2 was 0.08 (Figure 2D). The absence of correlation demonstrates that, beyond the greater number of MutL than MutS molecules on mismatches noted above, there is no regularity in the stoichiometric relationship between MutS and MutL on mismatches.
Taken together, these results show that a greater number of MutL molecules than MutS are involved in the mismatch repair reaction.
Number of MutL molecules on mismatches is independent of the intracellular concentration of MutL
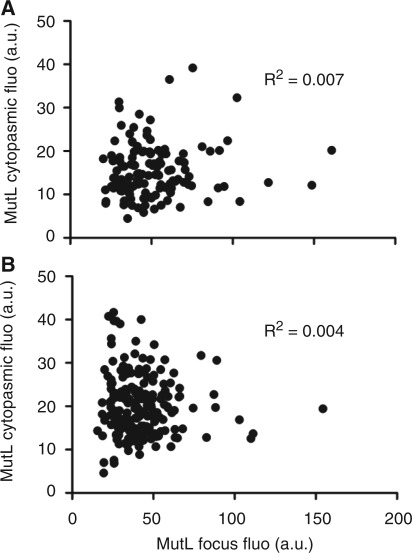
Since, we established that MutL assembles in greater numbers on mismatches than MutS, we wondered if this was determined by the intracellular concentration of MutL. To examine this possibility, we quantified the cytoplasmic fluorescence of eGFP-MutL and examined its correlation with the fluorescence intensity of MutL foci. No correlation between these fluorescence intensities was seen (Figure 3A). We conclude that the cytoplasmic concentration of MutL does not determine its amount assembled on mismatches.
Figure 3.
MutL amount on mismatches is independent of the intracellular concentration of MutL. No correlation between MutL focus fluorescence and cytoplasmic MutL fluorescence for (A) 131 foci without treatment (R2 = 0.007) and (B) 200 foci after 30-min treatment with rifamcipin (R2 = 0.004).
The absence of such correlation could result also from a limited life-time of the MutL foci (24). The disassembly of the MutL foci could affect the detection of any correlation between the amount of MutL in the cytoplasm and on mismatches. The disassembly of MutL foci depends on DNA replication (24). When replication initiation is blocked with rifampicin, the life-time of MutL foci is greatly increased, but their fluorescence does not increase (24). To test the possible effect of the disassembly of MutL foci on the correlation between the amount of MutL in the cytoplasm and on mismatches, we treated the cells with rifampicin in order to block the initiation of chromosome replication. There was also no correlation between the fluorescence intensity of the MutL foci and the cytoplasmic fluorescence in cells treated with rifampicin (Figure 3B).
Together, these results confirm that the fluorescence of MutL foci depends on the number of MutL molecules in the focus and not on its intracellular concentration or on the life-time of the focus.
Roadblocks on DNA reduce the number of MutL molecules assembled on mismatches
There are two possibilities as to why the mismatches lead to the accumulation of MutL. MutL could assemble along the DNA or it could form a complex that is not in contact with the DNA. These two possibilities can be distinguished by placing roadblocks on the DNA. If MutL assembles along the DNA, then such roadblocks would limit the number of MutL molecules in the foci. The decreased number of MutL molecules in the foci would decrease the foci fluorescence, or the frequency of foci, when the number of MutL molecules becomes insufficient for focus detection. If MutL forms a complex that does not make contact along the DNA, then such roadblocks would have no effect on the foci fluorescence or on the foci frequency.
The mutHE56A codes for a MutH protein that efficiently binds to GATC sequences in vitro without cleaving them (29). Due to this property, MutHE56A can be used as a roadblock on the GATC sequence proximal to a mismatch.
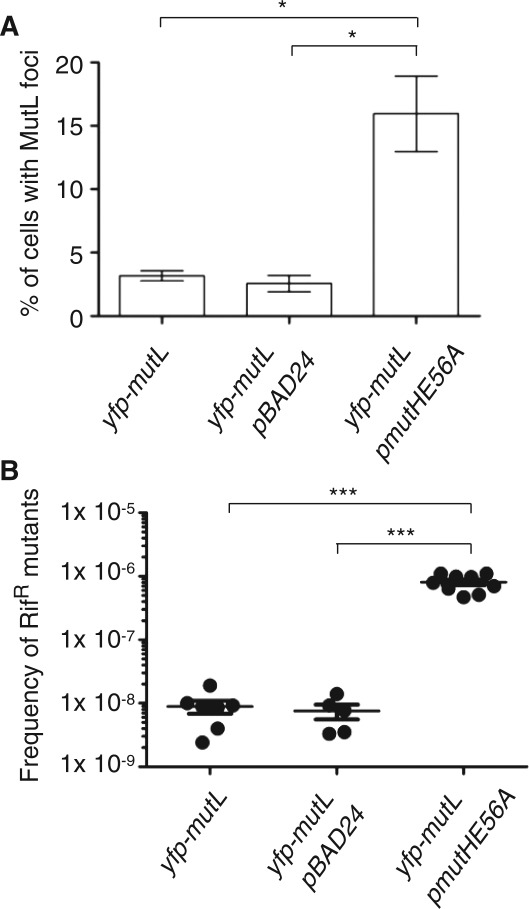
We first tested how the expression of MutHE56A affects the frequency of MutL foci in wild-type and in mutH cells. We showed previously that the frequency of fluorescent MutL foci corresponds closely to the mutation frequency of different strains examined (24). We found that the production of MutHE56A increases significantly the frequency of MutL foci in the wild-type cells compared to controls (Figure 4A). We confirmed this result by a classical mutagenesis assay, i.e. we measured the frequency of appearance of spontaneous mutations conferring resistance to rifampicin in strains expressing MutHE56A and in the control wild-type strains (Figure 4B). The results show that expression of MutHE56A increased spontaneous mutagenesis.
Figure 4.
Dominant negative effect of roadblock MutHE56A over wild-type MutH. MutHE56A expression in the wild-type cells increases significantly (A) the percentage of cells with at least 1 MutL focus (B) the frequency of rifampicin-resistant mutants, compared to controls (P < 0.05, Wilcoxon test).
We interpret this result to be a consequence of a dominant negative effect of the mutant over the wild-type protein expressed from its native chromosomal site. The binding of MutHE56A to GATC sequences likely competes with MutHwt rendering mismatch repair inefficient and explaining the increase in mutation frequency. In vivo dominant negative effect of MutHE56A is consistent with previous in vitro studies showing that MutHE56A binds GATC sequences as efficiently as MutHwt but is completely defective for the DNA incision (29).
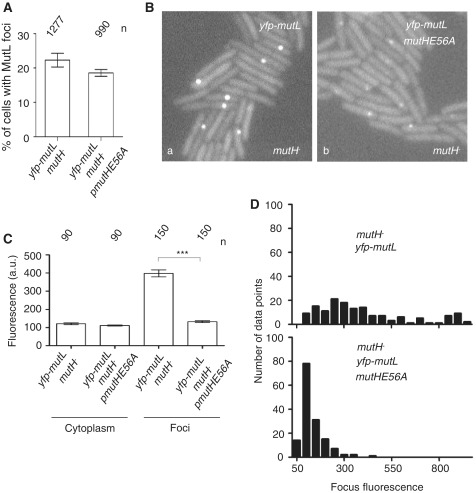
The expression of MutHE56A had no effect on the frequency of MutL foci in mutH cells compared to control cells (Figure 5A), arguing that these cells were still MMR deficient. On the other hand, the expression of MutHwt from a plasmid completely restored the MMR proficiency of the mutH cells. This was determined by measuring the frequency of YFP-MutL foci and the frequency of appearance of rifampicin resistant mutants (data not shown). These results are consistent with previous studies of mutation frequencies of mutH cells expressing MutHE56A or MutHwt (29).
Figure 5.
Roadblock on strand discrimination site reduces the accumulation of MutL on DNA mismatches. (A) Percentage of cells with at least 1 MutL focus in mutH cells is not affected by MutHE56A expression (P > 0.05, Wilcoxon test). (B) Fluorescent images of mutH cells expressing (a) fluorescent MutL alone or (b) fluorescent MutL and MutHE56A mutant. (C) The mean focus fluorescence for 150 foci from Ba and for 150 foci from Bb is significantly different (P < 0.0001, Wilcoxon test). The mean cytoplasmic fluorescence for 90 cells from B(a) and 90 cells from B(b) is not significantly different (P > 0.05, Wilcoxon test). (D) The histograms of MutL foci fluorescence for cells from B(a) and B(b). Error bars in A and C indicate the standard error of the mean. n in A and C indicates the number of cells/foci examined.
Finally, we examined the effect of MutHE56A expression on the amount of MutL associated with mismatches. We performed this experiment in mutH cells because they provide a large number of MutL foci, which is necessary for good statistics.
The fluorescence intensity of MutL foci was significantly lower in cells expressing MutHE56A compared to control cells that did not express this mutant protein (3-fold; P < 0.0001, Wilcoxon test) (Figure 5B–D). There was no difference in the intracellular concentration of MutL between the cells that expressed MutHE56A and those that did not, as judged by the fluorescence intensity of the cytoplasm (Figure 5C). This result suggests that MutL foci are less fluorescent as binding of MutHE56A to GATC prevents MutL from accumulating further along DNA. However, the decrease in MutL foci fluorescence and the increase in mutation frequency observed in the presence of MutHE56A could also be due to occluding the binding sites on MutL by overproducing one of its binding partners, in our case MutH. If this is so, we expect that the overproduction per se of wild-type or mutant MutH should have the same effect. However, our data with overproduction of wild-type MutH eliminate this hypothesis (Supplementary Figure S1).
Taken together, our results suggest that MutL assembles along the DNA from the mismatch site toward a proximal unmethylated GATC sequences.
DISCUSSION
MutL is a molecular matchmaker protein, which interacts with MutS, MutH and UvrD (2,6). MutL interaction with the MutS–mismatch complex triggers MutH endonuclease activity at a distal-strand discrimination DNA site (2,6). The nature of communication between these distal sites is not known. Several models based on cis and trans communication between these sites have been proposed (10–14). All of these models are based on in vitro studies and imply an equal stoichiometry of MutS and MutL proteins in the mismatch repair reaction. One hypothesis, that still awaits experimental evidence, suggests that the accumulation of MutL on DNA could allow the establishment of a physical interaction between the mismatch–MutS complex and the strand discrimination site (15). There are two predictions of this hypothesis that could be tested in vivo. The first is that the in vivo stoichiometry of mismatch repair involves more MutL than MutS molecules. The second is that a steric block on the DNA around a mismatch prevents the accumulation of MutL.
We used functional fluorescent MutS and MutL proteins to test these predictions. We found that MutS and MutL co-localize on mismatches in visible foci and that MutL foci are, on average, 2.7 times more intense than the co-localized MutS foci. These results indicate that the stoichiometry of the mismatch repair reaction in vivo involves more molecules of MutL than MutS. There is a caveat to this interpretation. Mismatch–MutS–MutL complexes that we are detecting are forming on mismatches that are unrepairable, due to mutH deficiency. It is possible that MutS and MutL stoichiometry on repairable mismatches is different from the one that we observe.
Our results on MutS and MutL stoichiometry on unrepaired mismatches are consistent with the in vitro DNA footprinting studies that show that MutS alone covers about 20 bp around mismatch, but that the addition of MutL increases considerably the size of the footprint (to 143 bp or more) (11,16). Our calculations, based on the resolved structure of MutS and on the model of the intact MutL (33–36), predict that one MutS dimer protects ∼20 bp while one MutL dimer covers ∼10 bp. Therefore, the extensive coverage of DNA seen in the presence of MutL is most likely due to the binding of multiple MutL proteins to the MutS-bound mismatch.
The second prediction of the model that suggests that MutL contacts the DNA while it accumulates around a mismatch is that a steric block on the GATC, proximal to the mismatch, would limit the amount of accumulated MutL. We used a mutant MutH protein, MutHE56A, which is able to bind to GATC sequences, but cannot cleave them. This protein was dominant negative when present in the cell and it reduced the amount of MutL present around mismatches. This is consistent with the idea that MutL contacts the DNA as it accumulates around a mismatch. This notion also suggests that MutL has at least two sites of where it can bind around a mismatch, to another MutL molecule and to the DNA. This predicts that the binding of MutL to the DNA could be cooperative. Indeed, it was shown that E. coli MutL, as well as its homologs, Mlh1-Pms1, bind DNA cooperatively in vitro (37,38).
On the account of the in vivo results presented here, and previous genetic and biochemical studies on MMR proteins, we propose the following model for in vivo MMR mechanism. Upon initial binding of MutS to a mismatch, MutS may diffuse and translocate along the DNA helix hence allowing additional copies of MutS to be loaded. Such repetitive MutS loading on mismatch is predicted by MMR models that involve movement of MMR proteins along the helix contour (14,17). Iterative binding of MutS to mismatches could explain the appearance of MutS foci in vivo. Binding of MutL to mismatch-bound MutS prevents MutS from sliding away as suggested by a study reporting an increased half-life of MutS–heteroduplex DNA complex upon MutL addition (10). Presumably, MutL does not affect MutS that is bound to mismatch but it interacts with MutS that slid away (14). MutL binding to DNA-bound MutS forms a ternary complex and establishes a nucleation point for further MutL accumulation on DNA surrounding the mismatch. Escherichia coli MutS tetramerization domain structure reveals that stable dimers but not tetramers are essential for DNA mismatch repair in vivo (39). A homodimeric MutS might be able to interact with two MutL dimers, which would lead to a ratio of MutL:MutS of 2 that is close to the observed values (2.1, for the combination of CFP-MutL and YFP-MutS, and 3.2 for the combination of YFP-MutS and CFP-MutL). However, our results show (data not shown) that, in all combinations, the fluorescence of MutS and MutL foci is highly variable and that the coefficient of variation of foci fluorescence is similar for differently labeled MutS and MutL. This suggests that not just one MutS and two MutL dimers accumulate on unrepaired mismatches but several MutS dimers and on average twice more dimers of MutL. We hypothesize that MutS–MutL–DNA filament-like structure is dynamic. A static filament might be inhibitory for strand discrimination as the binding of MutL to the DNA in a filament like manner could occlude the GATC sites similar as in MutS–MutL–DNA footprinting experiments (11,16).
Why may in vivo MMR require multiple MutL molecules? The average distance between two adjacent GATC sequences in E. coli genome is 256 bp. Except for runs of repeated mono-, di- and tri-nucleotides, the replicative DNA polymerase presumably makes errors randomly. Therefore, mismatches can be found anywhere in between two adjacent GATCs. So, the average distances between the mismatch and the proximal GATC are between 1 and 125 bp. Therefore, longer distances between mismatch and proximal GATC require accumulation of multiple copies of MutS and MutL proteins. In the light of our results, it would be interesting to test if the regions with low GATC abundance are more prone to mutations due to the requirement of higher numbers of the MutL protein, which appears to be in limiting quantity for replication fidelity control (40) as well as for recombination between non identical DNA sequences (41).
Our findings are also relevant for the understanding of the phenomenon of MutL saturation in vivo. MMR system has limited capacity to repair DNA mismatches: it saturates when the number of mismatches increases in the cell above a certain threshold. It was shown in several studies that MMR saturation is a result of MutL limitation, and not of limitation of MutS (18–22). However, the cause of MutL limitation was not identified. The simple possibility that MutL limitation is due to a lower amount of MutL in cell was excluded as both MutL and MutS are present in cells in equimolar concentrations (23). We found that treatments, which saturate MutL, do not lead to degradation of MutL that could lead to the deficit of functional MutL (Elez, M. and Radman, M., unpublished data). Therefore, in the light of our results, we propose that the deficit of MutL under conditions which increase the load of mismatches in cells could be due to engagement of more MutL than MutS proteins on the DNA mismatches. Unlike MutS, MutL associates with other members of the mismatch repair pathway in E. coli, the stoichiometry of which is unclear. This added involvement of MutL could also be an additional reason for MutL becoming saturating instead of MutS.
The MMR mechanism that involves more MutL accumulation relative to MutS observed in this study could be evolutionary conserved. For example, while this article was in revision, Kolodner's group published the article demonstrating the mechanism of MMR in yeast using the same in vivo approach as we did here (42). They reported that the mismatch recognition by Msh2–Msh6 directs formation of superstoichiometric Mlh1-Pms1 foci.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figure 1 and Supplementary Reference 43.
FUNDING
European Commission Cordis (FP7-HEALTH-F3-2010-241476) and from the Agence Nationale de la Rechearche (ANR-09-BLAN-0251); University Paris-Descartes (to M.E.) and Inserm Unit 1001 funds (to M.E.). Funding for the open access charge: European Commission Cordis (FP7-HEALTH-F3-2010-241476)
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Martin Marinus and Wei Yang for kindly providing bacterial strains and plasmids as indicated. We also acknowledge Alex Dajkovic, Lydia Robert, François Heslot and Mario Pende for critical reading of the manuscript. M.E., M.R. and I.M conceived the experiments. M.E. performed the experiments. M.E., M.R. and I.M. analyzed and interpreted the results, and wrote the paper.
REFERENCES
- 1.Radman M, Wagner BW, Glickman M, Messelson M. Progress in Environmental Mutagenesis. Elsevier, North-Holland, Amsterdam: 1980. [Google Scholar]
- 2.Kunkel TA, Erie DA. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 3.Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 4.Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech. Ageing Dev. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 6.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 7.Petit MA, Dimpfl J, Radman M, Echols H. Control of large chromosomal duplications in Escherichia coli by the mismatch repair system. Genetics. 1991;129:327–332. doi: 10.1093/genetics/129.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rayssiguier C, Thaler DS, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 9.Kolodner RD, Mendillo ML, Putnam CD. Coupling distant sites in DNA during DNA mismatch repair. Proc. Natl Acad. Sci. USA. 2007;104:12953–12954. doi: 10.1073/pnas.0705698104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schofield MJ, Nayak S, Scott TH, Du C, Hsieh P. Interaction of Escherichia coli MutS and MutL at a DNA mismatch. J. Biol. Chem. 2001;276:28291–28299. doi: 10.1074/jbc.M103148200. [DOI] [PubMed] [Google Scholar]
- 11.Selmane T, Schofield MJ, Nayak S, Du C, Hsieh P. Formation of a DNA mismatch repair complex mediated by ATP. J. Mol. Biol. 2003;334:949–965. doi: 10.1016/j.jmb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 12.Junop MS, Obmolova G, Rausch K, Hsieh P, Yang W. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell. 2001;7:1–12. doi: 10.1016/s1097-2765(01)00149-6. [DOI] [PubMed] [Google Scholar]
- 13.Allen DJ, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, Griffith JD. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol. Cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 15.Modrich P. DNA mismatch correction. Annu. Rev. Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- 16.Grilley M, Welsh KM, Su SS, Modrich P. Isolation and characterization of the Escherichia coli mutL gene product. J. Biol. Chem. 1989;264:1000–1004. [PubMed] [Google Scholar]
- 17.Blackwell LJ, Wang S, Modrich P. DNA chain length dependence of formation and dynamics of hMutSalpha.hMutLalpha.heteroduplex complexes. J. Biol. Chem. 2001;276:33233–33240. doi: 10.1074/jbc.M105076200. [DOI] [PubMed] [Google Scholar]
- 18.Schaaper RM, Radman M. The extreme mutator effect of Escherichia coli mutD5 results from saturation of mismatch repair by excessive DNA replication errors. EMBO J. 1989;8:3511–3516. doi: 10.1002/j.1460-2075.1989.tb08516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Damagnez V, Doutriaux MP, Radman M. Saturation of mismatch repair in the mutD5 mutator strain of Escherichia coli. J. Bacteriol. 1989;171:4494–4497. doi: 10.1128/jb.171.8.4494-4497.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macintyre G, Doiron KM, Cupples CG. The Vsr endonuclease of Escherichia coli: an efficient DNA repair enzyme and a potent mutagen. J. Bacteriol. 1997;179:6048–6052. doi: 10.1128/jb.179.19.6048-6052.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matic I, Babic A, Radman M. 2-aminopurine allows interspecies recombination by a reversible inactivation of the Escherichia coli mismatch repair system. J. Bacteriol. 2003;185:1459–1461. doi: 10.1128/JB.185.4.1459-1461.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Negishi K, Loakes D, Schaaper RM. Saturation of DNA mismatch repair and error catastrophe by a base analogue in Escherichia coli. Genetics. 2002;161:1363–1371. doi: 10.1093/genetics/161.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng G, Tsui HC, Winkler ME. Depletion of the cellular amounts of the MutS and MutH methyl-directed mismatch repair proteins in stationary-phase Escherichia coli K-12 cells. J. Bacteriol. 1996;178:2388–2396. doi: 10.1128/jb.178.8.2388-2396.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elez M, Murray AW, Bi LJ, Zhang XE, Matic I, Radman M. Seeing mutations in living cells. Curr. Biol. 2010;20:1432–1437. doi: 10.1016/j.cub.2010.06.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Natl Acad. Sci. USA. 2008;105:3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fontaine F, Stewart EJ, Lindner AB, Taddei F. Mutations in two global regulators lower individual mortality in Escherichia coli. Mol. Microbiol. 2008;67:2–14. doi: 10.1111/j.1365-2958.2007.05988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng G, Winkler ME. Single-step purifications of His6-MutH, His6-MutL and His6-MutS repair proteins of escherichia coli K-12. Biotechniques. 1995;19:956–965. [PubMed] [Google Scholar]
- 29.Junop MS, Yang W, Funchain P, Clendenin W, Miller JH. In vitro and in vivo studies of MutS, MutL and MutH mutants: correlation of mismatch repair and DNA recombination. DNA Repair. 2003;2:387–405. doi: 10.1016/s1568-7864(02)00245-8. [DOI] [PubMed] [Google Scholar]
- 30.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller JH. A Short Course in Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1992. [Google Scholar]
- 32.Smith BT, Grossman AD, Walker GC. Visualization of mismatch repair in bacterial cells. Mol. Cell. 2001;8:1197–1206. doi: 10.1016/s1097-2765(01)00402-6. [DOI] [PubMed] [Google Scholar]
- 33.Guarne A, Ramon-Maiques S, Wolff EM, Ghirlando R, Hu X, Miller JH, Yang W. Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair. EMBO J. 2004;23:4134–4145. doi: 10.1038/sj.emboj.7600412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ban C, Yang W. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell. 1998;95:541–552. doi: 10.1016/s0092-8674(00)81621-9. [DOI] [PubMed] [Google Scholar]
- 35.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 36.Natrajan G, Lamers MH, Enzlin JH, Winterwerp HH, Perrakis A, Sixma TK. Structures of Escherichia coli DNA mismatch repair enzyme MutS in complex with different mismatches: a common recognition mode for diverse substrates. Nucleic Acids Res. 2003;31:4814–4821. doi: 10.1093/nar/gkg677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall MC, Wang H, Erie DA, Kunkel TA. High affinity cooperative DNA binding by the yeast Mlh1-Pms1 heterodimer. J. Mol. Biol. 2001;312:637–647. doi: 10.1006/jmbi.2001.4958. [DOI] [PubMed] [Google Scholar]
- 38.Bende SM, Grafstrom RH. The DNA binding properties of the MutL protein isolated from Escherichia coli. Nucleic Acids Res. 1991;19:1549–1555. doi: 10.1093/nar/19.7.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendillo ML, Putnam CD, Kolodner RD. Escherichia coli MutS tetramerization domain structure reveals that stable dimers but not tetramers are essential for DNA mismatch repair in vivo. J. Biol. Chem. 2007;282:16345–16354. doi: 10.1074/jbc.M700858200. [DOI] [PubMed] [Google Scholar]
- 40.Galan JC, Turrientes MC, Baquero MR, Rodriguez-Alcayna M, Martinez-Amado J, Martinez JL, Baquero F. Mutation rate is reduced by increased dosage of mutL gene in Escherichia coli K-12. FEMS Microbiol. Lett. 2007;275:263–269. doi: 10.1111/j.1574-6968.2007.00902.x. [DOI] [PubMed] [Google Scholar]
- 41.Elez M, Radman M, Matic I. The frequency and structure of recombinant products is determined by the cellular level of MutL. Proc. Natl Acad. Sci. USA. 2007;104:8935–8940. doi: 10.1073/pnas.0610149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 147:1040–1053. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Sys. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.