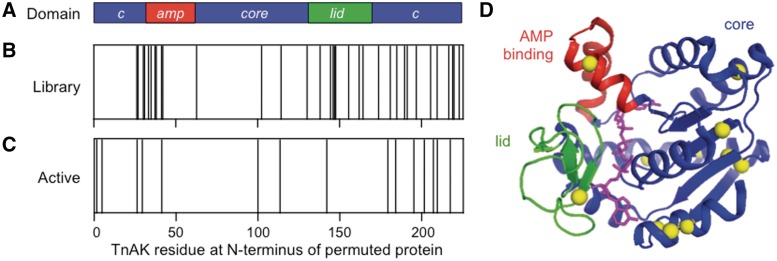
Figure 4.
Sequences of permuted TnAK from the unselected and selected libraries. (A) Color coding of the core (blue) AMP binding (red) and lid (green) domains in TnAK sequence helps the reader map the location of new N- and C-termini in the permuted variants. (B) The TnAK residues encoded by the first codon in unselected adk are indicated with a line, as well as (C) selected variants that complement E. coli CV2. (D) The TnAK residues encoded by the first codon in each functional variant (yellow spheres) are mapped onto the structure of Bacillus subtilis AK (36), which displays 48% sequence identity with TnAK. The substrate analog P1,P5-di(adenosine-5) pentaphosphate is shown in magenta. The image was created using PyMOL.