Figure 3. Ligand binding.
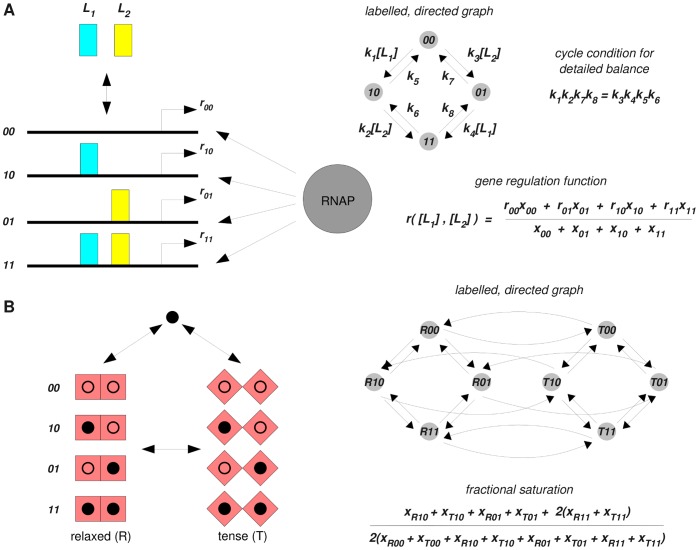
A. Gene regulation, with two transcription factors (TFs), L
1 and L
2, binding to a promoter. A labelled, directed graph can be constructed as described in the text, with the microstates being denoted here by the bitstrings 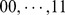 . In this example there are no changes of conformational state in the scaffold, as would be the case if there was DNA looping or displacement of nucleosomes. The rate of mRNA transcription by RNA polymerase (RNAP) is assumed to depend on the pattern of TF binding as shown and the overall rate is calculated as an average over the probabilities of finding the promoter in each of the patterns. Assuming the system is at an equilibrium, x, these probabilities are given by the ratios,
. In this example there are no changes of conformational state in the scaffold, as would be the case if there was DNA looping or displacement of nucleosomes. The rate of mRNA transcription by RNA polymerase (RNAP) is assumed to depend on the pattern of TF binding as shown and the overall rate is calculated as an average over the probabilities of finding the promoter in each of the patterns. Assuming the system is at an equilibrium, x, these probabilities are given by the ratios,  , which can be calculated using the second elimination formula in (4). B. An allosteric homodimeric protein is shown in two conformational states, relaxed (R) and tense (T). The labelled, directed graph has both conformational state changes in the scaffold as well as ligand binding and unbinding. The microstates are denoted here
, which can be calculated using the second elimination formula in (4). B. An allosteric homodimeric protein is shown in two conformational states, relaxed (R) and tense (T). The labelled, directed graph has both conformational state changes in the scaffold as well as ligand binding and unbinding. The microstates are denoted here 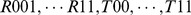 . Labels have been omitted for clarity. For allosteric enzymes, the overall rate of product formation is usually assumed to be proportional to the fraction of sites that are bound by ligand (fractional saturation), as shown. This can be calculated as described in Supporting Information S1.
. Labels have been omitted for clarity. For allosteric enzymes, the overall rate of product formation is usually assumed to be proportional to the fraction of sites that are bound by ligand (fractional saturation), as shown. This can be calculated as described in Supporting Information S1.