Table 2.
Mammalian oxidized base-specific DNA glycosylases and their preferred substrates. Oxidative modifications are indicated in red.
| OGG1 | NTH1 | NEIL1 | NEIL2 |
|---|---|---|---|
(i) 8-oxoG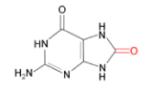
|
(i) Thymine glycol
|
(i) 5-OHU (and 5-OHC)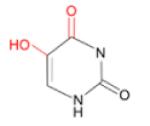
|
(i) Guanidinohydantoin
|
(ii) FapyG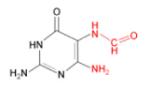
|
(ii) 5-OHU
|
(ii) FapyA (and FapyG)
|
(ii) 5-OHU
|
(iii) 5-OHC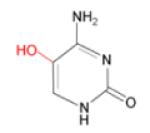
|
(iii) 8-oxoG
|
(iii) Dihydrouracil (DHU)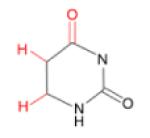
|
|
(iv) 5-formy1U
|
(iv) Thymine glycol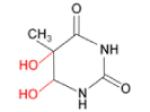
|
