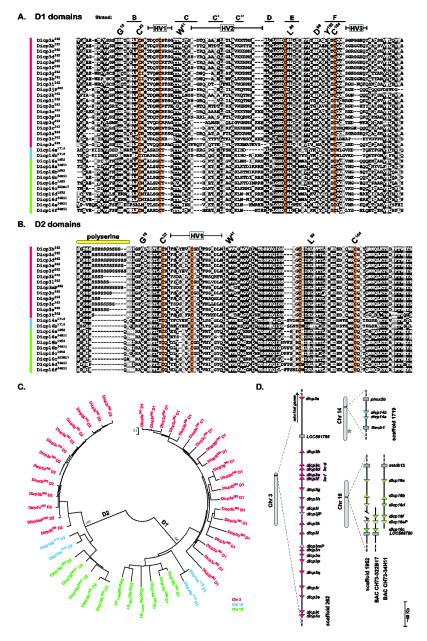
Fig. 1. DICP D1 and D2 domains encoded by genomic sequences.
DICP Ig domains were organized into (A) D1 and (B) D2 domains and aligned. Positions that are 70% or greater identical are shaded in black and those that are structurally related are shaded in gray. Protein symbols are shown on the left with the genomic source (superscript) indicated as an allele designation (scaffold 262, scaffold 1719, scaffold 1952, BAC CH73-34H11 or BAC CH73-322B17). Conserved residues characteristic of immunoglobulin domains are indicated by the IMGT numbering system above the alignments [37]. Hypervariable regions (HV) and predicted Ig strand locations (of D1) also are indicated. A variable length polyserine stretch is evident at the amino terminal region of the D2 domain. Conserved cysteines are shaded with orange. (C) The DICP D1 and D2 domains shown in (A) and (B) were aligned and a neighbor joining tree constructed. (D) The relative chromosomal positions of DICP D1 domains are shown. Triangles represent a single DICP D1 domain; transcriptional orientation is indicated. Predicted pseudogenes are indicated by lighter shades of color and gray rectangles represent linked (non-DICP) genes. The dicp3c and dicp3d D1 domains and the dicp3e and dicp3f D1 domains are predicted to be encoded in single genes (see Supplemental Materials and Methods). BACs were employed to refine the sequence in scaffold 1952. The star on chromosome 14 indicates the location of a NITR gene cluster.