Abstract
Protein arginine (Arg) methylation serves an important functional role in eukaryotic cells, and typically occurs in domains consisting of multiple Arg in close proximity. Localization of methylarginine (MA) within Arg-rich domains poses a challenge for mass spectrometry (MS)-based methods; the peptides are highly-charged under electrospray ionization (ESI), which limits the number of sequence-informative products produced by collision induced dissociation (CID), and loss of the labile methylation moieties during CID precludes effective fragmentation of the peptide backbone. Here the fragmentation behavior of Arg-rich peptides was investigated comprehensively using electron transfer dissociation (ETD) and CID for both methylated and unmodified glycine-/Arg-rich peptides (GAR), derived from residues 679-695 of human nucleolin, which contains methylation motifs that are widely-represented in biological systems. ETD produced abundant information for sequencing and MA localization, whereas CID failed to provide credible identification for any available charge state (z=2-4). Nevertheless, CID produced characteristic neutral losses that can be employed to distinguish among different types of MA, as suggested by previous works and confirmed here with product ion scans of high accuracy/resolution by an LTQ/Orbitrap. To analyze MA-peptides in relatively complex mixtures, a method was developed that employs nano-LC coupled to alternating CID/ETD for peptide sequencing and MA localization/characterization, and an Orbitrap for accurate precursor measurement and relative quantification of MA-peptide stoichiometries. As proof of concept, GAR-peptides methylated in vitro by protein arginine N-methyltransferases PRMT1 and PRMT7 were analyzed. It was observed that PRMT1 generated a number of monomethylated (MMA) and asymmetric-dimethylated peptides, while PRMT7 produced predominantly MMA peptides and some symmetric-dimethylated peptides. This approach and the results may advance understanding of the actions of PRMTs and the functional significance of Arg methylation patterns.
1. Introduction
Protein arginine (Arg) methylations are prevalent in eucaryotes, and represent an important class of post-translational modifications (PTM) that can alter essential biological processes such as signal transduction, cellular proliferation, transcriptional processing, protein-protein interactions, and splicing of mRNA [1-3]. Protein Arg-methylation is also associated with numerous diseases such as HIV, cancer, metabolic disorders and neurodevelopmental disorders [4-12]. Methylarginines (MA) in proteins exist as monomethylarginine (MMA), asymmetric dimethylarginine (aDMA), or symmetric dimethylarginine (sDMA). Arg methylations frequently occur in eukaryotic cells in Arg-rich domains, such as repetitive RGG-, RXR- and DRG box motifs [13-15].
Determination of the locations and types of MA residues in proteins, and the quantification of methylation stoichiometries, is essential to understand the regulatory, physiological, and pathological functions associated with protein methylation. Conventional methods for the detection and characterization of MA residues include the incorporation of radioactive methyl groups, amino acid analysis, and Edman sequencing [16-18]. Shortcomings of these methods include the inability to determine which specific Arg residues are methylated, the necessity for laborious separation, or very low analytical throughput [19]. Identification of MA residues by matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry, using either MS or MS/MS analysis, has been reported [19-21] . However, methods based upon MALDI-TOF cannot be applied to complex samples, and may require prior knowledge of the target MA protein/peptide.
Alternatively, liquid chromatography coupled to electrospray ionization tandem mass spectrometry (LC/ESI-MS/MS), which employs collision induced dissociation (CID) for peptide fragmentation, has been used to achieve peptide separation and identification of MA residues in relatively complex samples such as protein digest mixtures [21-26]. However, analysis of MA-peptides by MS/MS-based approaches employing ESI for ionization and CID for fragmentation can be challenging for several reasons. First, Arg methylations often occur in domains of multiple Arg residues in close proximity that contain adjacent Arg box motifs [13-15]. Because trypsin seldom cleaves at the C-terminus of MA residues [27], tryptic digestion of MA proteins often results in peptides containing multiple internal MA residues. Second, MA residues are rather basic, and thus amenable to protonation in the gas phase. As a result, MA peptides are usually highly charged under ESI conditions (≥3 charges), and often provide only limited sequence-informative b and y ions during CID [28], perhaps because the positively charged internal MA impedes the mobile proton on the peptide backbone [29]. This lack of sequence-informative ions renders it difficult to sequence MA-rich peptides or to localize the methylated Arg residues by CID. Third, MA moieties in peptides, particularly dimethylarginine (DMA) residues, are labile under CID conditions. Thus neutral losses such as dimethylamine (dma), dimethylcarbodiimide (dmc), or monomethylamine (mma) predominate over peptide backbone fragmentation. As a result, CID spectra tend to be dominated by a small number of fragments resulting from neutral losses and contain few sequence-informative ions [22, 23], and information on peptide sequence and methylation position cannot be obtained (cf. Fig. 3). It was observed previously that CID of the multiply-charged MA peptide VAAR*GR*GR*GMGR*GK (where *=DMA) produced spectra consisting predominantly of several ions resulting from neutral loss and containing very few b and y ions [23]. Other studies showed similar trends for peptides containing multiple MA residues when fragmented by CID [30]. Finally, recent studies suggest that Arg methylation may parallel phosphorylation in its level of complexity, in terms of the locations at which methylations can occur, and the possible stoichiometries at each location [13]. This complexity further compounds the difficulties in the analysis of MA peptides.
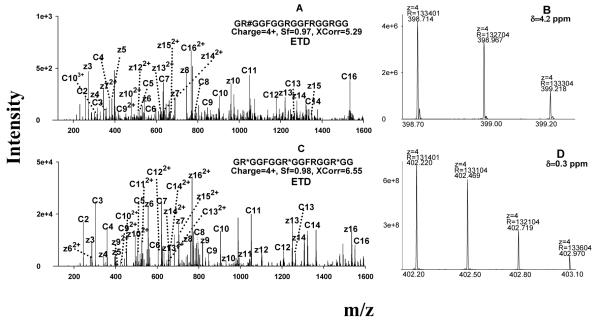
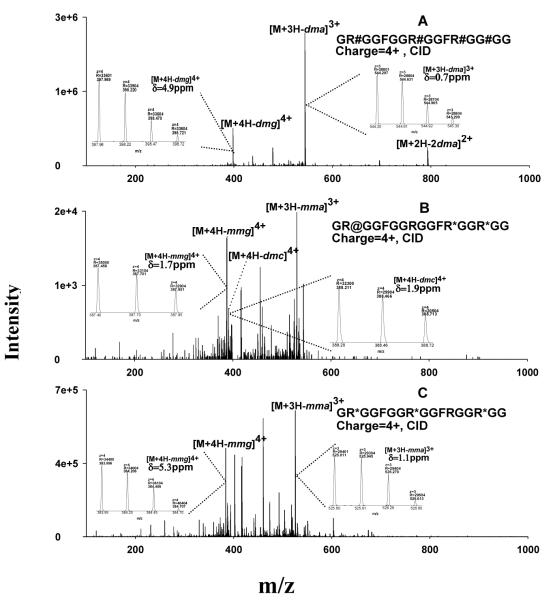
Figure 3.
Representative MS spectra for the identification of the MA-peptides produced by in vitro methylation systems. (A) ETD spectrum of a PRMT1-produced peptide identified as GR#GGFGGRGGFRGGRGG, where #=aDMA; the symmetry of the DMA was determined separately by CID; (B) confirmation of the identification of the peptide GR#GGFGGRGGFRGGRGG by accurate precursor measurement using the Orbitrap; (C) ETD spectrum of a PRMT7-produced peptide identified as GR*GGFGGR*GGFRGGR*GG, where *=MMA; (D) confirmation of the identification of the peptide as GR*GGFGGR*GGFRGGR*GG using the Orbitrap data. Abbreviation: δ, mass error in ppm.
The technical challenges described above may explain why few studies describe the analysis of sequence, localization, and characterization of MA-peptides by ESI-MS/MS, despite the biological importance of Arg methylations. Therefore, a robust method for accurate localization of MA residues and quantification of methylation stoichiometries in Arg-rich peptides would represent a significant analytical advancement that could lead to new biological insights into the mechanisms of protein methylation and its significance in biological and pathological processes.
Here we describe the development of an analytical approach based on LC/ESI-MS that provides improved identification and characterization of MA-containing peptides within Arg-rich domains. Our approach was to employ electron transfer dissociation (ETD), which is a fragmentation technology developed recently by Hunt and Coon [31], in order to identify highly-charged, Arg-rich methylated peptides and to localize the sites of methylation. ETD converts multiply charged peptide cations into radicals through reaction with radical gaseous anions (e.g. fluoranthene) via an electron transfer process. The peptide cation radicals then undergo fragmentation of the N-Cα bond as a result of their poor stability [32-35]. The resulting fragment ions that contain the N- and C termini of the peptides are termed c and z ions, respectively. Although ETD usually does not fragment doubly charged peptide ions efficiently, it does provide fairly uniform cleavage of peptide precursors of higher charge states (≥3) [31, 36]. A recent study by Coon and coworkers suggested that the fragmentation efficiency and resulting sequence coverage by ETD is approximately proportional to the charge density (i.e. number of charges/number of residues) of the precursor peptide ions [28]. Because Arg methylations frequently occur in Arg-rich domains that contain multiple closely-spaced Arg residues, it could be expected that both the charge states and charge densities of such peptide ions would be high under ESI conditions; this should provide optimal peptide backbone fragmentation under ETD. Moreover, ETD is particularly well suited for the characterization of peptides containing labile modifications, because ETD often leaves them intact during peptide bond dissociation [33, 37]. Preservation of methylation moieties during peptide bond dissociation would provide abundant information on the location of the modification.
The experimental system employed a biologically-relevant Arg-rich peptide, denoted GAR (Glycine- and Arginine- Rich peptide) [38] (GRGGFGGRGGFRGGRGG), which was synthesized both with and without specific MA modifications, or subjected to methylation by in vitro enzymatic methylation systems. The GAR peptide comprises residues 679-695 of human nucleolin (NP_005372), which is one of the most abundant nucleolar proteins of vertebrate cells. Nucleolin is involved in biological processes such as ribosome biogenesis, transcription, embryogenesis, and cell proliferation and growth, as well as in pathological conditions such as specific systemic autoimmune disorders [4]. Methylation of the GAR domain is essential for the function and localization of nucleolin [39], and is the target of a potent pseudopeptide inhibitor of both HIV entry [40] and cancer [41]. The methylation motifs contained in the GAR peptide are common to many naturally occurring methylated proteins [13-15]. Furthermore, despite the important biological and pathophysiological significance of Arg methylations as a family of post-translational regulatory modifications, methylation patterns generated by prevalent enzymatic systems such as the protein arginine N-methyltransferases (PRMTs) are not well characterized. Two PRMTs, PRMT1 and PRMT7, were employed to investigate the patterns of peptide methylation produced enzymatically.
In terms of the experimental approach, ETD was used to sequence the methylated GAR peptides and to determine the positions of methylations. CID fragmentation was used to characterize the type of MA derivatization (i.e. MMA, sDMA or aDMA). In order to ensure high-confidence identification, an Orbitrap analyzer, which provides high accuracy/resolution for the measurement of m/z [42-44], was used to obtain accurate m/z determinations of the peptide precursor ions and CID fragment ions, as well as to perform relative quantification of the stoichiometries of the enzymatically-methylated peptides.
Experimental
Materials
HPLC grade methanol, acetonitrile, and water were from B&J (Muskegon, MI). Formic acid was from Fluka (Buchs, Switzerland). Phenylmethanesulfonyl fluoride (PMSF), benzamidine and S-adenosylmethionine were obtained from Sigma (St. Louis). Caffeine, MAFA peptide, and Ultramark for Obitrap calibration were from Thermo Scientific (San Jose, CA). Microcon centrifugal filters were obtained from Millipore (Billerica, MA). Two synthetic MA-rich peptides, the unmodified GAR peptide (GRGGFGGRGGFRGGRGG) and the per-aDMA GAR peptide (GR#GGFGGR#GGFR#GGR#GG, where #=aDMA), were synthesized by the laboratory of Sandor Pongor (International Center for Genetic Engineering and Biotechnology, Trieste, Italy). The amino acid sequence of the GAR peptide corresponds to the residues 679-695 of human nucleolin (NP_005372). The C-termini of both peptides were amidated.
In vitro methylation of the GAR peptide by PRMT1 and PRMT7
Two protein arginine methyltransferases, PRMT1 and PRMT7, were employed to produce MA-peptides in vitro using the unmodified GAR peptide as the substrate. Both enzymes are recombinant GST-fusion proteins; PRMT1 was the same enzyme preparation used in a previous work [7] and PRMT7 was generously provided by Drs. Laurie Read and John Fisk (Department of Microbiology, University at Buffalo School of Medicine). The unmodified GAR peptide was dissolved in water at a concentration of 80 μg/mL. Either 10 μg/mL PRMT1 or 4.3 μg/mL PRMT7 was added along with 65 mM Tris (pH 8), 1 mM PMSF, 10 μM benzamidine and 100 μM S-adenosylmethionine. The mixtures were then incubated for 20 hours at 23 °C. The reaction solutions were loaded onto Microcon centrifugal membrane (30 kDa MWCO) filter assemblies to remove the enzymes from the peptides. Following 10 minutes of centrifugation at 12,000 g, approximately 85% of the reaction solution containing MA-peptides was recovered as filtrate.
Nano-LC/MS analysis and identification of MA-peptides
A nano-LC system consisting of a Spark Endurance autosampler (Emmen, Holland) and four Eksigent direct-flow capillary/nano LC pumps (Dublin, CA) that were powered by pressurized nitrogen (100 p.s.i) were used for the separation of MA-peptides. The nano-LC flow path was lab-made and included a Vici 10-port low-dead-volume valve (Houston, TX). A lab-made dynamic flow nano-spray interface was used to couple the nano-LC system to a Thermo Scientific LTQ/ETD or to a LTQ/Orbitrap (San Jose, CA).
Mobile phase A consisted of 0.1% formic acid in 2% acetonitrile and mobile phase B was 0.1% formic acid in 84% acetonitrile. For analysis, MA-peptides were loaded onto the reversed-phase (RP) peptide trap (5 mm x 300 μm I.D., Agilent, Palo Alto, CA) at a flow rate of 10 μL/min and then washed with mobile phase A for 3 minutes. The trap was then switched online, and a lab-packed reversed-phase nano C-18 column was employed for separation. The column consisted of 3 μm reversed-phase particles packed in a 25 cm long, 360 μm O.D. and 75μm I.D. fused silica capillary terminated with a non-coated taperedtip of 2-3 μm diameter. The flow rate was 250 nL/min and the gradient consisted of: (i) a linear increase from 0 to 7% B over 5 min; (ii) an increase from 7 to 19% B over 55 min; (iii) an increase from 19 to 90% B in 1 min, and finally (iv) isocratic at 90% B for 10 min.
For LTQ/ETD, the MS was working in data-dependent mode; one scan cycle was comprised of an MS1 scan (m/z range from 300-2000 with a target value of3×104) followed by six sequential dependent MS2 scans (the target value was 1×104 and the maximum injection time was 250 ms). The first, third, and fifth MS2 scans were CID fragmentations of the first, second, and third most-abundant precursors, respectively, presenting in the MS1 spectrum. The second, fourth, and sixth MS2 scans were ETD fragmentations corresponding to the same group of precursors. For CID, the activation time was 30 ms, the isolation width was 1.5 amu, the normalized activation energy was 35%, and the activation q was 0.25. For ETD, a mixture of ultrapure helium and nitrogen (25% Helium and 75%, purity >99.995%) was used as the reaction gas. To obtain efficient fragmentation, a high target value for fluoranthene anions (2×106) was used; the ETD reaction time was set at 120 ms and the isolation width was 2 amu; supplemental activation, which uses a short CID activation process to dissociate the charge-stripped precursors, was employed to enhance the fragmentation efficiency for doubly-charged precursors. For Orbitrap analysis, one scan cycle included an MS1 scan (m/z 300-2000) at a resolution of 100,000 followed by three MS2 scans (resolution at 30,000) to fragment the three most abundant precursors found in the MS1 spectrum. The target values for MS1 and MS2 by Orbitrap were, respectively, 2×106 and 2×105. To ensure mass accuracy, the Orbitrap analyzer was calibrated on the same days that the experiments were performed. For both LTQ/ETD and Orbitrap experiments, dynamic exclusion was used with one repeat counts, 35s repeat duration, and 100s exclusion duration.
We employed an offline precursor matching approach to obtain accurate m/z and charge states of the MA-peptides putatively identified by LTQ/ETD. The same sample was analyzed by nano-LC/LTQ/Orbitrap immediately following the nano-LC/LTQ/ETD analysis, using the same chromatographic system, gradient conditions and nano-spray interface. The precursor m/z and retention times of the peptides that were putatively identified using LTQ/ETD were matched against data from the LTQ/Orbitrap, using an m/z window threshold of +/− 0.5 amu and a retention time window threshold of 0.2 min. Owing to the excellent run-to-run reproducibility of the nano-LC system, the matches were facile.
Spectra generated by ETD were processed using BioWorks software (3.3.1, Thermo Scientific), which incorporates the SEQUEST algorithm, to find the best matching sequences. Briefly, the charge states of the precursors of ETD spectra were assigned by the Charger program (Thermo Scientific) and then searched against a database containing the sequence of the GAR peptide. Differential modifications of MMA (+14.0156 Da) and DMA (+28.0313 Da) on Arg residues, and a static modification of C-terminal amidation (−0.9840 Da) were considered. Mass tolerances were 1.4 and 1.0 amu, respectively for the precursor and fragments. The resulting identifications were first filtered using the scores of Xcorr (Cross-correlation score; 2+ precursor ≥2.5, 3+ ≥ 3.2, 4+ ≥ 3.8) and then using the Sf scores (final score, ≥ 0.8). In cases where more than one methylation pattern resulted for one ETD spectrum, final confirmation of the most probable assignment was obtained by manual inspection of the spectrum for c and z ions. For putatively identified MA-peptides, the charge states and accurate m/z of precursors were obtained by the Orbitrap, and any identification with incorrect charge state assignment or precursor mass error larger than 10 ppm was eliminated.
Relative quantification of MA-peptides
To calculate the stoichiometries of the MA-peptides produced by the in vitro methylation systems, the extracting ion currents (XIC) of the precursors obtained by the Orbitrap were utilized for relative quantification. For each identified MA-peptide, the XICs were extracted in a narrow m/z window (±0.01 amu) around the monoisotopic m/z for each available charge state. The area-under-curve (AUC) for each precursor at each charge state was calculated using Qualbrowser (Thermo Scientific). The relative abundance (RA) of a MA-peptide (peptide A) was calculated using the following formula:
| [1] |
Results and Discussion
Ionization and fragmentation behaviors of methylated and non-methylated Arg-rich GAR peptides
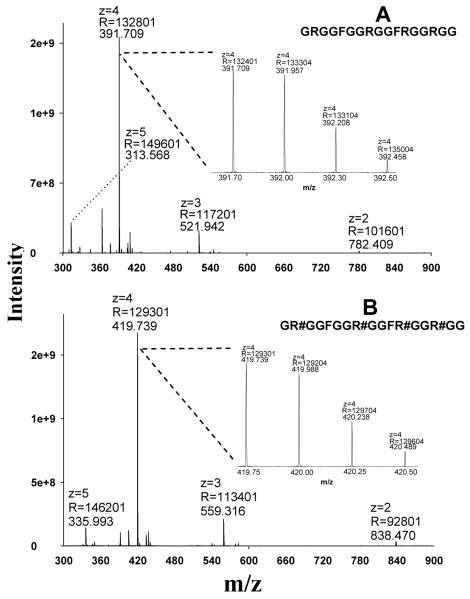
We and others have shown that the CID reactivity, fragmentation behavior and the amount of obtainable sequence-informative product ions of a peptide ion are heavily dependent upon the charge state of the target peptide [29, 45-47]. Therefore, it was essential to investigate the ionization behavior of MA-rich peptides prior to studying fragmentation. The GAR-domain peptide of nucleolin, which contains prevalent Arg-rich consensus domains that are methylated in biological systems, was synthesized both in unmodified form and with asymmetric dimethylations (aDMA) on all Arg residues (per-aDMA GAR). Using ESI and an Orbitrap analyzer, the charge state distribution for both peptides was determined readily (Fig. 1). The unmodified GAR peptide was predominantly quadruply-charged, with much less abundant double-, triple- and quintuple-charge state ions (Fig. 1A). Because MA residues would be considered less basic than unmodified Arg residues in biochemical processes [13], it might be expected that the charge state distribution of the per-aDMA GAR peptide would shift toward lower charge states. However, it was observed that the per-aDMA GAR peptide exhibited a charge state distribution almost identical to its unmodified cognate (Fig. 1B), suggesting that MA residues possess affinity for protons similar to unmodified Arg residues during the ESI process.
Fig. 1.
The charge state distributions of the GAR peptides under positive ESI, as obtained using an Orbitrap analyzer. (A) The MS1 spectrum of the unmodified peptide GRGGFGGRGGFRGGRGG; (B) the MS1 spectrum of the per-aDMA peptide GR#GGFGGR#GGFR#GGR#GG, where #=aDMA. Abbreviations: z, charge state; R, resolution.
In order to characterize the fragmentation behavior of the unmodified- and per-aDMA GAR peptides, both were subjected to CID and ETD activation. The CID spectrum of the predominant, quadruply-charged unmodified GAR peptide yielded a number of fragments arising from internal cleavages [19], as well as neutral losses and fragments produced via other undesirable pathways (Fig. 2A). However, very few sequence-informative b and y ions were observed. This observation can be explained by the ‘mobile proton’ model [29, 48]: for effective peptide backbone cleavage under CID conditions, an ionizing proton must be transferred to positions along the length of the peptide, weakening the amide bond at those positions and facilitating their cleavage. According to this model, the multiple fixed protons localized on the internal Arg residues of the highly-charged GAR peptide would impede the movement of the mobile proton, thus preventing uniform cleavage of the peptide backbone, which is necessary to obtain abundant sequence information.
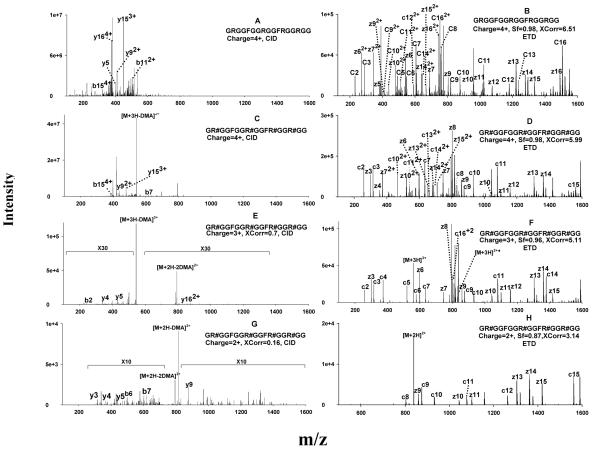
Figure 2.
Representative CID and ETD spectra of the unmodified (GRGGFGGRGGFRGGRGG) and per-aDMA GAR peptides (GR#GGFGGR#GGFR#GGR#GG, where #=aDMA) using precursors of different charge states. (A) CID of the quadruply-charged unmodified GAR peptide; (B) ETD of the quadruply-charged unmodified GAR peptide; (C) CID of the quadruply-charged per-aDMA GAR peptide; (D) ETD of the quadruply-charged per-aDMA GAR peptide; (E) CID of the triply-charged per-aDMA GAR peptide; (F) ETD of the triply-charged per-aDMA GAR peptide; (G) CID of doubly-charged per-aDMA GAR peptide; (H) ETD of the doubly-charged per-aDMA GAR peptide. CID did not result in any confident identification for the GAR peptides, even with low-charge-state precursors. Abbreviations: Xcorr, cross-correlation score; Sf, final score (ETD only). The criteria for a confident identification was Xcorr>2 if z=2, Xcorr >2.8 if z=3, Xcorr >4 if z=4; for ETD, Sf >0.8 was also required.
Similarly, the CID of the quadruply-charged per-aDMA GAR peptide also produced a limited number of b and y ions. Moreover, because the MA moieties are labile during CID process, the CID spectrum was dominated by a fragment corresponding to a facile cleavage of dma from the precursor (Fig. 2C). Due to the limited sequence information provided by these spectra, sequencing by SEQUEST did not result in any identification. Clearly, CID was not suitable for sequencing these highly-charged MA-peptides.
In contrast, ETD provided uniform dissociations of the peptide backbone of the same precursors and provided highly abundant sequence information. Nearly-complete c and z ion ladders were obtained for both the unmodified and per-aDMA GAR peptides (Fig. 2B and 2D), enabling highly confident sequencing of the peptides, as expressed by the high scores of Xcorr and Sf achieved. Because ETD is not sensitive to side chain chemistry, cleavage of the labile methylation moieties was not observed in the product ion spectrum of the per-aDMA GAR peptide, and the methyl moieties were well-preserved on c and z ions (Fig. 2D). Clearly, ETD is an ideal technique for the analysis of MA- and Arg- rich peptides, as neither the high charge states nor the labile methylations limited the production of abundant sequence information.
It can be observed from Fig. 1 that doubly- (2+) and triply- (3+) charged ions were also generated during ESI, although at very low abundance (<2% and <10% of the intensity of the 4+ species, respectively). Because CID can often produce relatively abundant sequence information for doubly- and triply- charged peptides [29, 48], it may be possible to use CID to sequence MA-rich peptides, provided the concentration of these peptides is sufficiently high in the sample so that the 2+ or 3+ precursors have the necessary abundance to provide a reasonable yield of fragments. To test this possibility, we investigated the CID fragmentation behavior of the 2+ and 3+ precursors of per-aDMA GAR using a peptide solution of high concentration (80 μg/mL). Nevertheless, surprisingly little sequence information were obtained; the CID spectra for both charge states were dominated by ion(s) corresponding to neutral losses of methylation moieties, and limited numbers and low abundances of b and y ions (Fig. 2E and 2G). SEQUEST identification using these spectra yielded very poor Xcorr scores (0.7 and 0.16 for 3+ and 2+ charged precursors, respectively), which were far below prevailing criteria for confidence (e.g. Xcorr>2.0 for 2+ and >2.5 for 3+). There are several possible explanations for this observation. First, because of the lability of the MA moieties, CID may promote initial elimination of the methylation moiety, thus largely precluding further fragmentation of the peptide backbone. Second, the peptide contains four internal MA residues, which are highly basic in the gas phase, as discussed above. As a result, the two (for the 2+ precursor) or three (for the 3+ precursor) ionizing protons are most likely localized on the MA residues, rather than on the less basic N-terminus. According to the ‘mobile proton’ model, a mobile proton associated with the N-terminus is essential for even peptide backbone cleavage [29, 48], and the lack of mobile proton on these 2+ and 3+ precursors would result in a very limited number of sequence-informative b and y ions in the CID spectra.
In contrast, ETD fragmentation of both the 2+ and 3+ precursors produced significantly more sequence information (Fig. 2F and 2H). Interestingly, ETD of the 2+ precursor showed mostly “long” c and z ions (i.e., fragments containing more than half of the total residues of the peptide) rather than a complete set of both ions (Fig. 2H). Because the 2+ precursor was charge-reduced in the ETD process, and one precursor ion cannot dissociate into both c and z ions when only one net positive charge is available, the number of c and z ion species obtained from a 2+ precursor is significantly less than the number obtained from more highly charged precursors. Nevertheless, the fragments in Fig. 2H represent 11 inter-residue cleavages out of a total of 16 inter-residue bonds in the peptide, which provided satisfactory sequence coverage and consequently a highly confident identification (Xcorr=3.14 and Sf=0.87). As a result of the use of supplemental activation, as well as a high anion intensity for ETD (cf. the Experimental section), ETD fragmentation efficiency for the 2+ precursor was fairly high (Fig. 2H) and thus probably contributed to the high SEQUEST peptide scores obtained.
Confident determination of methylation patterns and relative quantification of the stoichiometries of methylarginines on MA-GAR peptides produced in vitro by enzymatic methylation
The majority of Arg methylations in eukaryotic cells appear to be performed by members of the protein arginine N-methyltransferase (PRMT) family [13]. Specific PRMT enzymes could be selective for particular Arg locations or motifs, and produce distinct methylation patterns [4, 13]. Therefore, determination of the types and positions of methylations produced by these enzymes on common Arg- rich motifs, such as are present in the GAR peptide, is essential to understand their mechanisms of action and role in Arg methylation in biological systems. To date, there exists no reliable method for the identification and characterization of these enzymatically-produced methylations, largely as a result of the inability of CID to analyze Arg-rich peptides, as demonstrated above. Furthermore, given the perplexing position preference of enzymatic methylation [13], determination of the exact localization of each methylation among closely spaced Arg residues is clearly very challenging. As demonstrated with the synthesized per-aDMA GAR peptide (above), ETD was able to provide a nearly complete ladder of c and z ions, and also preserved the labile Arg methylations of highly-charged precursors, which is essential for accurate localization of MA residues. Therefore, ETD was investigated for the analysis of enzymatically methylated GAR peptides.
The products of two representative in vitro enzymatic methylation systems, employing PRMT1 and PRMT7, were analyzed using nano-LC coupled with ETD fragmentation and Orbitrap analysis. Although it would be possible to conduct the analysis by direct infusion of the reaction mixtures into the MS system, nano-LC was employed to eliminate the possible effects of sample buffers and reaction reagents on the ionization of the MA-peptides. Furthermore, because there are four Arg residues in the peptide, isomeric MA-peptides may be produced that have the same mass adduction (by MA) but are methylated at different positions. Chromatographic resolution of these isomers was essential for the identification and localization MA positions on these peptides (cf. Fig. 4).
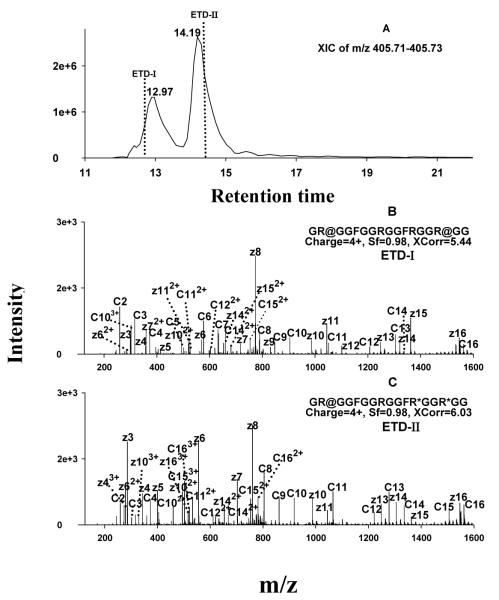
Figure 4.
Typical ETD spectra and chromatogram for unambiguous identification of isomeric MA-peptides. (A) XIC extracted within the narrow m/z range of 405.71-405.73; the vertical dashed lines denote the times when ETD fragmentations were performed. (B) ETD spectrum acquired at the time of ETD-I, which enabled the identification of the earlier-eluting isomer as GR@GGFGGRGGFRGGR@GG (where @=sDMA; the symmetry of the DMA was determined in parallel by CID); (C) ETD spectrum acquired at the time of ETD-II, which identified the later-eluting isomer as GR@GGFGGRGGFR*GGR*GG (where *=MMA).
a. Determination of the types and locations of Arg methylations on enzymatically produced MA-GAR peptides
As observed with the synthesized methylated peptides (above), GAR peptides methylated by both enzymatic systems were also predominantly quadruply-charged under positive ESI (spectra not shown). ETD fragmentation of these ions produced nearly complete ladders of complementary c and z ions, and thus enabled highly confident peptide identification, as well as accurate localization of the MA-residues. In those cases in which not all Arg residues were methylated, localization of the methylations by SEQUEST was confounded because of the close proximity of the four Arg residues in the GAR peptide. In fact, SEQUEST often provided several possible assignments of methylation patterns from a single ETD spectrum, with some of the assignments having similar scores. In such cases, manual examination of the c and z ions was necessary in order to determine the specific modification sites, and the MA-peptides tentatively identified by ETD were examined further using high accuracy precursor measurements.
For proteomic research employing MS, high mass accuracy measurement of peptide precursor ions assists considerably in the elimination of non-obvious false positives [49]. Therefore, a high resolution/accuracy Orbitrap analyzer [42-44] was used to measure the m/z of precursor ions of the MA-peptides identified by ETD. Because an integrated Orbitrap-ETD instrument was not available, samples were analyzed sequentially on LTQ-ETD and LTQ-Orbitrap instruments, and offline precursor matching was employed (Experimental section). Because of the excellent run-to-run reproducibility of the nano-LC system, offline matches were facile (chromatograms not shown). The accurate m/z and charge state information obtained for the putative MA-peptides using the Orbitrap was used to filter MA-peptides in the ETD data; any identification in which the precursor had incorrect charge state assignments or the mass error was greater than 10 ppm was considered a false positive identification.
It was observed that PRMT1 and PRMT7 produced various patterns of GAR peptide methylations. Under the reaction conditions used here, the major methylation type catalyzed by both enzymes was MMA, but lower abundance DMA products were also produced (Table 1). Integration of the high-accuracy m/z data from the Orbitrap with the ETD data reduced considerably the number of putative identifications; of the 17 MA-peptides provisionally identified by ETD, 5 were eliminated because the precursors did not fall within the 10 ppm mass error criteria. It is important to note that all of these false-positive identifications had high scores. This finding emphasizes the importance of highly accurate measurement of the m/z of precursor ions in order to eliminate false-positive identifications that otherwise appear acceptable. All peptide identifications that passed the acceptance criteria are shown in Table 1.
Table 1.
a Nano-LC/CID/ETD-Orbitrap identification and relative quantification of methylarginine residues on a nucleolin peptide produced by in vitro methylation
| Sequence | m/z (4+) |
b Relative abundance (%) |
c Sf/Xcorr |
d Mass error (ppm) |
|---|---|---|---|---|
| Standard peptides | ||||
| GRGGFGGRGGFRGGRGG | 391.709 | - | 0.98/6.51 | 0.25 |
| e GR#GGFGGR#GGFR#GGR#GG | 419.739 | - | 0.98/5.99 | 0.02 |
| Methylation products of PRMT7 | ||||
| GRGGFGGRGGFRGGRGG | 391.709 | 1.0 | 0.97/6.05 | 1.50 |
| GRGGFGGRGGFRGGR*GG | 395.212 | 12.6 | 0.98/6.55 | 0.02 |
| GR*GGFGGRGGFRGGR*GG | 398.715 | 25.1 | 0.98/6.77 | 1.69 |
| GR@GGFGGRGGFRGGR@GG | 405.725 | 0.4 | 0.98/5.44 | 2.58 |
| GR@GGFGGRGGFR*GGR*GG | 405.722 | 0.3 | 0.98/6.03 | 4.16 |
| GRGGFGGRGGFR*GGR*GG | 398.717 | 4.5 | 0.98/5.93 | 2.35 |
| GR*GGFGGR*GGFRGGR*GG | 402.220 | 56.1 | 0.98/6.55 | 0.28 |
| Methylation products of PRMT1 | ||||
| GRGGFGGRGGFRGGRGG | 391.709 | 85.2 | 0.98/6.32 | 1.91 |
| GR#GGFGGR#GGFR#GGR#GG | 419.739 | 2.1 | 0.98/5.4 | 0.81 |
| GR#GGFGGRGGFRGGRGG | 398.714 | 2.5 | 0.97/5.29 | 4.16 |
| GR*GGFGGRGGFR*GGRGG | 398.715 | 1.3 | 0.98/5.63 | 2.50 |
| GR*GGFGGR*GGFRGGRGG | 398.715 | 1.2 | 0.98/5.66 | 2.39 |
| GRGGFGGR*GGFRGGRGG | 395.212 | 2.1 | 0.98/5.82 | 0.48 |
| GR*GGFGGRGGFRGGRGG | 395.211 | 5.6 | 0.98/6.44 | 2.15 |
A synthesized peptide corresponding to residues 679-695 of human nucleolin (NP_005372), was used as the substrate for both PRMT1 and PRMT7.
The relative abundance of each peptide was normalized against the total summed abundance of peptides in the sample.
The Sf and Xcorr are two scores that evaluate the confidence of SEQUEST identification of an ETD spectrum. Values of Sf>0.80 and Xcorr >4 for z=4 were considered high confidence.
The mass error for the precursor ions (observed m/z vs. theoretical m/z) measured by Orbitrap
#=aDMA, @=sDMA and *=MMA
To investigate whether some MA-peptides were generated via sample preparation artifacts, as previously reported by Qin and co-workers [24], we also analyzed negative control samples in which the GAR peptide was incubated in the reaction mixture lacking enzyme. No MA-peptides were identified in the negative control samples (data not shown).
ETD spectra used for the identification of two representative MA-peptides, one dimethylated by PRMT1 and the other monomethylated by PRMT7, are shown in Fig. 3A and Fig. 3C. Accurate m/z- and charge state information obtained with the Orbitrap confirmed the identifications (Fig. 3B and Fig. 3D). Not surprisingly, CID provided very poor sequence information for these MA-peptides, for the reasons discussed above (spectra not shown).
Sufficient chromatographic separation was found to be essential for the identification of isomeric MA-peptides, which are expected to occur fairly frequently by enzymatic methylation within domains rich in closely-spaced Arg residues. Isomeric MA peptides possess the same peptide backbone and m/z, but differ in the position of the methylations; the polarity of such isomeric MA-peptides is close, and therefore they elute chromatographically in close proximity. If isomeric peptides were not separated, the ETD spectra would contain mixed c and z ions from multiple isomers, which could result in incorrect localization, low matching scores, or even a failure of identification, as we observed in preliminary experiments (not shown). Therefore, we optimized a shallow gradient separation procedure to resolve isomeric peptides (cf. Experimental section). Fig. 4 shows an example in which the combination of sufficient nano-LC separation and ETD fragmentation enabled unambiguous identification of isomeric MA peptides produced enzymatically by PRMT7. Two quadruply-charged isomers, having an observed m/z of 405.723, were resolved (Fig. 4A). ETD fragmentation performed within each of the retention time windows of the two isomers provided nearly-complete ladders of sequence-informative fragments and thus enabled accurate determination of the types and positions of methylations on both (Fig. 4B and 4C). The presence of dimethylated z3-z15 and c3-c14 (Fig. 4B), and the fact that both c15 and z16 carry two dimethyl groups, clearly indicates that the MA-peptide that eluted at 13.0 min is dimethylated on the two Arg residues closest to the C- and N-termini. Dimethylated c2-c4 ions and monomethylated z3-z8 ions were observed for the peptide that eluted at 14.2 min. In addition, z11 and z13 carry two MMA, and c9-c13 carry one MMA and one DMA. These observations permitted the locations of the methylations to be determined unambiguously, as shown in Fig. 4C. The DMA residues on both peptides were subsequently identified as sDMA residues by CID, as described below.
b. Identification of methylation types via characteristic neutral losses under CID combined with high-resolution product ion scan
The ability to identify different types of methylations, and particularly the discrimination of aDMA and sDMA, is important because of reported differences in biological actions [15, 23]. Because the methylation moieties were observed to remain intact during ETD fragmentation, it is not feasible to differentiate aDMA and sDMA by ETD. However, although CID was not suitable for the sequencing of Arg-rich MA-peptides, previous studies demonstrated that peptides containing different types of methylations produced characteristic and specific neutral loss fragments [19, 22, 23] , which are summarized in Table 2. Previous assignments of the characteristic neutral losses of MA-peptides were performed with low-resolution/accuracy MS analyzers such as ion traps and quadrupoles. The use of an Orbitrap analyzer for this task is advantageous because the accurate m/z and charge state information provided would not only enable a highly confident examination/confirmation of these proposed neutral loss species, but also would aid in the interpretation of fragment spectra. Therefore, the Orbitrap was employed to perform a close inspection of the CID fragmentation spectra of MA-GAR-peptides. Representative spectra are shown in Fig. 5. For the synthesized per-aDMA peptide, the most abundant fragments were confirmed to result from neutral loss of dma and dmg, with less than 5 ppm mass error (Fig. 5A). For a MA-GAR peptide produced enzymatically by PRMT7, the CID spectrum contained fragments characteristic of both sDMA and MMA, but not aDMA, suggesting that the DMA near the N-terminus is an sDMA (Fig. 5B). In Fig. 5C, the presence of high-abundance neutral losses of mmg and mma indicates that the MA-peptide contains only MMA, which agrees with the ETD results (Fig 3C).
Table 2.
Summary of characteristic CID neutral losses from different types of methylarginines as proposed by previous studies [19, 22, 23]
| Neutral loss | a Mass (Da) | b Methylation type |
|---|---|---|
| Dimethylamine (dma) | 45.058 | aDMA |
| Dimethylguanidine (dmg) | 87.087 | aDMA or sDMA |
| Dimethylcarbodiimide (dmc) | 70.053 | sDMA |
| Monomethylamine (mma) | 31.042 | MMA or sDMA |
| Monomethylguanidine (mmg) | 73.064 | MMA |
The values are all for monoisotopic mass;
Abbreviations: aDMA, asymmetric dimethylarginine; sDMA, symmetric dimethylarginine; MMA, monomethylarginine.
Figure 5.
Typical high resolution/accuracy CID spectra of GAR peptides bearing different types of methylations, which were acquired by LTQ/Orbitrap. (A) CID spectrum of GR#GGFGGR#GGFR#GGR#GG (#=aDMA); (B) CID spectrum of the PRMT7-produced peptide GR@GGFGGRGGFR*GGR*GG (where @=sDMA and *=MMA; (C) CID spectrum of the PRMT7-produced peptide GR*GGFGGR*GGFRGGR*GG. Abbreviations: dma=dimethylamine, dmg=dimethylguanidine, dmc=dimethylcarbodiimide, mma=monomethylamine, mmg=monomethylguanidine.
CID of MA-peptides not only permits discrimination of sDMA vs. aDMA, but also may aid sequencing by providing information supplementary to that obtained by ETD. For example, in the event that the c and z ions obtained from an Arg-rich peptide are insufficient to discriminate between the alternative possibilities of (i) MMA on two Arg residues in close proximity, or (ii) DMA on a single Arg residue, the type-specific neutral losses obtainable with CID would eliminate this ambiguity. This capability of CID could be particularly useful if the abundance of the MA-peptide was relatively low, which is frequently the case for many biological samples. Under such conditions, ETD may not be able to produce a sufficiently complete sequence ladder.
It is interesting to note that the types of methylation products produced by PRMT7 have been debated previously; Miranda et al. observed that PRMT7 produced only MMA [50], whereas Lee and coworker showed that PRMT7 catalyzed only the formation of sDMA [51]. Here we observed that PRMT7 produced both MMA and sDMA on the GAR peptide (Table 1). As for PRMT1, we observed that both MMA and aDMA were generated. Relative quantification by Orbitrap analysis revealed that the stoichiometry of these methylations was significantly different for the two enzymes. (Table 1 and below).
c. Quantification of the Arg methylation stoichiometry produced by PRMT enzymes
The stoichiometry of PRMT methylation products is of interest for understanding the biological role of this family of enzymes. Our strategy to quantify MA stoichiometry was to utilize the precursor ion signals measured by the Orbitrap analyzer, which provides a wide dynamic range and high selectivity [52]. The AUC of the extracting ion current (XIC) for all charge states of a given MA-peptide were summed, and the relative quantity of the MA-peptide was calculated as described in the Experimental section. To ensure specificity, the XICs were extracted within a narrow m/z window (0.02 amu). Table 1 shows the relative abundances calculated for all identified MA-peptides generated enzymatically by PRMT1 and PRMT7. The isomeric MA-peptides listed were all resolved chromatographically, and the AUCs for each isomer were obtained separately. From the observation that the unmodified peptide substrate represented only 1% of the total peptides in the PRMT7 reaction mixture but 85.2% of the PRMT1 reaction mixture, it can be concluded that the PRMT7 used in this study was significantly more efficient than PRMT1 in the in vitro system employed. Also it can be observed (Table 1) that for both enzymes, the majority of the Arg methylation products are MMA. For PRMT7 in particular, sDMA-peptides account for less than 1% of the total MA-peptides produced, and the predominant products bear MMA on two or three Arg residues.
It should be noted that this quantification approach is based on the assumption that all forms of the GAR peptide (methylated and non-methylated) exhibit similar ionization efficiencies. This assumption appears to be reasonable due to the observation that the ionization efficiencies of the unmodified- and per-aDMA peptides were very similar (cf. Fig. 1). It is also important to note that because one form of methylation at a specific residue location often exists among different MA-peptides, the calculated relative abundances are per MA-peptide, but not necessarily per Arg residue position. Finally, this quantification strategy may not be readily adaptable in highly complex systems such as tryptic digests of biological samples, where differential ion suppression effects could compromise quantification accuracy. To address this problem, it may be feasible to incorporate internal standards for quantification in biological samples, and this possibility will be investigated in ongoing studies.
Conclusions
The confidence of peptide sequencing by database searching relies heavily on the extent and quality of the sequence information generated by MS/MS events [42]. Because of their intrinsically high charge states under ESI and the lability of methyl moieties, the sequencing of methylated Arg-rich peptides is difficult when CID is used as the means of fragmentation. Although the details of the dissociation mechanism by ETD have not been elucidated fully, it appears that ETD neither relies upon the mobile protons to dissociate peptides, nor is sensitive to side chain chemistry [31, 36]. Employing ETD fragmentation to investigate methylated Arg-rich peptides that contain methylation motifs widely observed in eukaryotic biological systems, the current work demonstrated that ETD fragmentation produced abundant sequence information for all available charge states, and permitted highly confident sequencing and accurate localization of methylations. CID, in contrast, provided few sequence-informative fragments for these peptides. However, CID can assist in the determination of the types of methylations, and particularly in the discrimination between aDMA and sDMA, as suggested by previous work [19, 22, 23] and confirmed here by the high-resolution/accuracy product ion scans produced by the LTQ/Oribitrap. Because of these complementary capabilities, combining alternating ETD and CID in a single run provided the data necessary for the comprehensive identification of MA-peptides.
The extent to which this strategy will permit elucidation of Arg methylation patterns in the non-arginine-rich motifs that exist biologically remains to be determined. However, missed tryptic cleavages at the C terminus of MA is a well recognized phenomenon, and this produces peptide charge states > +2. Furthermore, methylations in other peptide motifs are likely to be labile under CID, as was observed for the peptides investigated here. Thus there is confidence that the advantages of ETD in this combined ETD/CID strategy will provide the ability to elucidate methylation patterns in a wider range of biologically-occurring motifs than represented in the peptides analyzed here.
The products of two enzymatic methylation systems were investigated as proof-of-principle for application of this approach to biological systems. Following nano-LC separation, the MA-peptides generated by PRMT1 and PRMT7 were sequenced and the methylations were localized using ETD. The types of methylations were determined by using CID fragmentation that interleaved with ETD in a single run. To confirm identifications, high resolution/accuracy Orbitrap data was employed to eliminate false-positives. It was observed that PRMT1 produced both MMA- and aDMA- peptides with relatively low efficiency, as the majority of the substrate was unmodified in the reaction system used here. In contrast, the PRMT7 preparation used here converted approximately 85% of the substrate to MA-peptides, and generated both MMA and sDMA. Relative quantification of the stoichiometry of these products by Orbitrap analysis revealed that the vast majority of the MA-peptides produced by PRMT7 were MMA-peptides.
The approach developed here, employing nano-LC separation, interleaved CID and ETD, and parallel Orbitrap analysis, permitted facile sequencing, accurate methylation localization, and relative quantification of methylation stoichiometry for enzymatically-produced peptides that contain biologically-relevant Arg-rich motifs. This capability will provide valuable insight into the biological roles and mechanisms of action of these enzymes. Furthermore, this strategy may be adapted to the analysis of methylated proteins from biological samples after digestion of target proteins with the appropriate enzymes, which is currently on-going in our lab.
Acknowledgements
This work was supported by CH3 BioSystems LLC and a University at Buffalo Center of Advanced Technology (CAT) award from the New York State Office of Science Technology and Academic Research. This work was also partially supported by the Center of Protein Therapeutics grant of University at Buffalo. The LTQ/ETD system was obtained by Shared Instrumentation Grant S10-RR021221 from the National Center for Research Resources, National Institutes of Health (USA). We thank Drs. Laurie Read and John Fisk of the Department of Microbiology, University at Buffalo School of Medicine for their generous gift of the PRMT7 enzyme and the laboratory of Dr. Sandor Pongor at the International Centre for Genetic Engineering and Biotechnology (Trieste, Italy) for peptide synthesis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Boisvert FM, Cote J, Boulanger MC, Richard S. A proteomic analysis of arginine-methylated protein complexes. Molecular & Cellular Proteomics. 2003;2:1319–1330. doi: 10.1074/mcp.M300088-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, David M. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. 2001;104:731–741. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- 3.Dolzhanskaya N, Merz G, Aletta JM, Denman RB. Methylation regulates the intracellular protein-protein and protein-RNA interactions of FMRP. J. Cell Sci. 2006;119:1933–1946. doi: 10.1242/jcs.02882. [DOI] [PubMed] [Google Scholar]
- 4.Zahariev S, Guarnaccia C, Zanuttin F, Pintar A, Esposito G, Maravic G, Krust B, Hovanessian AG, Pongor S. Efficient synthesis and comparative studies of the arginine and Nomega,Nomega-dimethylarginine forms of the human nucleolin glycine/arginine rich domain. J Pept Sci. 2005;11:17–28. doi: 10.1002/psc.577. [DOI] [PubMed] [Google Scholar]
- 5.Nisole S, Said EA, Mische C, Prevost MC, Krust B, Bouvet P, Bianco A, Briand JP, Hovanessian AG. The anti-HIV pentameric pseudopeptide HB-19 binds the C-terminal end of nucleolin and prevents anchorage of virus particles in the plasma membrane of target cells. J. Biol. Chem. 2002;277:20877–20886. doi: 10.1074/jbc.M110024200. [DOI] [PubMed] [Google Scholar]
- 6.Thompson PR, Fast W. Histone citrullination by protein arginine deiminase: is arginine methylation a green light or a roadblock? ACS Chem Biol. 2006;1:433–441. doi: 10.1021/cb6002306. [DOI] [PubMed] [Google Scholar]
- 7.Pelletier M, Xu Y, Wang X, Zahariev S, Pongor S, Aletta JM, Read LK. Arginine methylation of a mitochondrial guide RNA binding protein from Trypanosoma brucei. Mol. Biochem. Parasitol. 2001;118:49–59. doi: 10.1016/s0166-6851(01)00367-x. [DOI] [PubMed] [Google Scholar]
- 8.Mostaqul Huq MD, Gupta P, Wei LN. Post-translational modifications of nuclear co-repressor RIP140: a therapeutic target for metabolic diseases. Curr. Med. Chem. 2008;15:386–392. doi: 10.2174/092986708783497382. [DOI] [PubMed] [Google Scholar]
- 9.Stetler A, Winograd C, Sayegh J, Cheever A, Patton E, Zhang X, Clarke S, Ceman S. Identification and characterization of the methyl arginines in the fragile X mental retardation protein Fmrp. Hum. Mol. Genet. 2006;15:87–96. doi: 10.1093/hmg/ddi429. [DOI] [PubMed] [Google Scholar]
- 10.Yong J, Wan L, Dreyfuss G. Why do cells need an assembly machine for RNA-protein complexes? Trends Cell Biol. 2004;14:226–232. doi: 10.1016/j.tcb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Iacovides DC, O’Shea CC, Oses-Prieto J, Burlingame A, McCormick F. Critical role for arginine methylation in adenovirus-infected cells. J. Virol. 2007;81:13209–13217. doi: 10.1128/JVI.01415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aletta JM, Hu JC. Protein arginine methylation in health and disease. Biotechnol. Annu. Rev. 2008;14:203–224. doi: 10.1016/S1387-2656(08)00008-2. [DOI] [PubMed] [Google Scholar]
- 13.McBride AE, Silver PA. State of the Arg: Protein methylation at arginine comes of age. Cell. 2001;106:5–8. doi: 10.1016/s0092-8674(01)00423-8. [DOI] [PubMed] [Google Scholar]
- 14.Patton WF. Detection technologies in proteome analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;771:3–31. doi: 10.1016/s1570-0232(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 15.McBride AE, Weiss VH, Kim HK, Hogle JM, Silver PA. Analysis of the yeast arginine methyltransferase Hmt1p/Rmt1p and its in vivo function - Cofactor binding and substrate interactions. J. Biol. Chem. 2000;275:3128–3136. doi: 10.1074/jbc.275.5.3128. [DOI] [PubMed] [Google Scholar]
- 16.Smith JJ, Rucknagel KP, Schierhorn A, Tang J, Nemeth A, Linder M, Herschman HR, Wahle E. Unusual sites of arginine methylation in Poly(A)-binding protein II and in vitro methylation by protein arginine methyltransferases PRMT1 and PRMT3. J. Biol. Chem. 1999;274:13229–13234. doi: 10.1074/jbc.274.19.13229. [DOI] [PubMed] [Google Scholar]
- 17.Klein S, Carroll JA, Chen Y, Henry MF, Henry PA, Ortonowski IE, Pintucci G, Beavis RC, Burgess WH, Rifkin DB. Biochemical analysis of the arginine methylation of high molecular weight fibroblast growth factor-2. J. Biol. Chem. 2000;275:3150–3157. doi: 10.1074/jbc.275.5.3150. [DOI] [PubMed] [Google Scholar]
- 18.Liu Q, Dreyfuss G. In-Vivo and in-Vitro Arginine Methylation of Rna-Binding Proteins. Mol. Cell. Biol. 1995;15:2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gehrig PM, Hunziker PE, Zahariev S, Pongor S. Fragmentation pathways of N(G)-methylated and unmodified arginine residues in peptides studied by ESI-MS/MS and MALDI-MS. J. Am. Soc. Mass Spectrom. 2004;15:142–149. doi: 10.1016/j.jasms.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Chiou YY, Lin WJ, Fu SL, Lin CH. Direct mass-spectrometric identification of Arg296 and Arg299 as the methylation sites of hnRNP K protein for methyltransferase PRMT1. Protein J. 2007;26:87–93. doi: 10.1007/s10930-006-9049-9. [DOI] [PubMed] [Google Scholar]
- 21.Zou Y, Wang Y. Tandem mass spectrometry for the examination of the posttranslational modifications of high-mobility group A1 proteins: symmetric and asymmetric dimethylation of Arg25 in HMGA1a protein. Biochemistry (Mosc) 2005;44:6293–6301. doi: 10.1021/bi0475525. [DOI] [PubMed] [Google Scholar]
- 22.Ong SE, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat Methods. 2004;1:119–126. doi: 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- 23.Brame CJ, Moran MF, McBroom-Cerajewski LD. A mass spectrometry based method for distinguishing between symmetrically and asymmetrically dimethylated arginine residues. Rapid Commun. Mass Spectrom. 2004;18:877–881. doi: 10.1002/rcm.1421. [DOI] [PubMed] [Google Scholar]
- 24.Jung SY, Li Y, Wang Y, Chen Y, Zhao Y, Qin J. Complications in the assignment of 14 and 28 Da mass shift detected by mass spectrometry as in vivo methylation from endogenous proteins. Anal. Chem. 2008;80:1721–1729. doi: 10.1021/ac7021025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou Y, Wang Y. Mass spectrometric analysis of high-mobility group proteins and their post-translational modifications in normal and cancerous human breast tissues. J Proteome Res. 2007;6:2304–2314. doi: 10.1021/pr070072q. [DOI] [PubMed] [Google Scholar]
- 26.Zou Y, Webb K, Perna AD, Zhang Q, Clarke S, Wang Y. A mass spectrometric study on the in vitro methylation of HMGA1a and HMGA1b proteins by PRMTs: methylation specificity, the effect of binding to AT-rich duplex DNA, and the effect of C-terminal phosphorylation. Biochemistry (Mosc) 2007;46:7896–7906. doi: 10.1021/bi6024897. [DOI] [PubMed] [Google Scholar]
- 27.Hayakawa M, Hosogi Y, Takiguchi H, Saito S, Shiroza T, Shibata Y, Hiratsuka K, Kiyama-Kishikawa M, Abiko Y. Evaluation of the electroosmotic medium pump system for preparative disk gel electrophoresis. Anal. Biochem. 2001;288:168–175. doi: 10.1006/abio.2000.4901. [DOI] [PubMed] [Google Scholar]
- 28.Good DM, Wirtala M, McAlister GC, Coon JJ. Performance characteristics of electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 2007;6:1942–1951. doi: 10.1074/mcp.M700073-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Qu J, Straubinger RM. Improved sensitivity for quantification of proteins using triply charged cleavable isotope-coded affinity tag peptides. Rapid Commun. Mass Spectrom. 2005;19:2857–2864. doi: 10.1002/rcm.2138. [DOI] [PubMed] [Google Scholar]
- 30.Rappsilber J, Friesen WJ, Paushkin S, Dreyfuss G, Mann M. Detection of arginine dimethylated peptides by parallel precursor ion scanning mass spectrometry in positive ion mode. Anal. Chem. 2003;75:3107–3114. doi: 10.1021/ac026283q. [DOI] [PubMed] [Google Scholar]
- 31.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JE, Coon JJ. Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Anal. Chem. 2007;79:477–485. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coon JJ, Shabanowitz J, Hunt DF, Syka JE. Electron transfer dissociation of peptide anions. J. Am. Soc. Mass Spectrom. 2005;16:880–882. doi: 10.1016/j.jasms.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 34.Gunawardena HP, He M, Chrisman PA, Pitteri SJ, Hogan JM, Hodges BD, McLuckey SA. Electron transfer versus proton transfer in gas-phase ion/ion reactions of polyprotonated peptides. J. Am. Chem. Soc. 2005;127:12627–12639. doi: 10.1021/ja0526057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coon JJ, Ueberheide B, Syka JE, Dryhurst DD, Ausio J, Shabanowitz J, Hunt DF. Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McAlister GC, Phanstiel D, Good DM, Berggren WT, Coon JJ. Implementation of electron-transfer dissociation on a hybrid linear ion trap-orbitrap mass spectrometer. Anal. Chem. 2007;79:3525–3534. doi: 10.1021/ac070020k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JEP, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2006;1764:1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raman B, Guarnaccia C, Nadassy K, Zakhariev S, Pintar A, Zanuttin F, Frigyes D, Acatrinei C, Vindigni A, Pongor G, Pongor S. N(omega)-arginine dimethylation modulates the interaction between a Gly/Arg-rich peptide from human nucleolin and nucleic acids. Nucleic Acids Res. 2001;29:3377–3384. doi: 10.1093/nar/29.16.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghisolfi L, Joseph G, Amalric F, Erard M. The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J. Biol. Chem. 1992;267:2955–2959. [PubMed] [Google Scholar]
- 40.Krust B, Vienet R, Cardona A, Rougeot C, Jacotot E, Callebaut C, Guichard G, Briand JP, Grognet JM, Hovanessian AG, Edelman L. The anti-HIV pentameric pseudopeptide HB-19 is preferentially taken up in vivo by lymphoid organs where it forms a complex with nucleolin. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14090–14095. doi: 10.1073/pnas.221467298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu X, Hamhouyia F, Thomas SD, Burke TJ, Girvan AC, McGregor WG, Trent JO, Miller DM, Bates PJ. Inhibition of DNA replication and induction of S phase cell cycle arrest by G-rich oligonucleotides. J. Biol. Chem. 2001;276:43221–43230. doi: 10.1074/jbc.M104446200. [DOI] [PubMed] [Google Scholar]
- 42.Yates JR, Cociorva D, Liao L, Zabrouskov V. Performance of a linear ion trap-Orbitrap hybrid for peptide analysis. Anal. Chem. 2006;78:493–500. doi: 10.1021/ac0514624. [DOI] [PubMed] [Google Scholar]
- 43.Scherl A, Shaffer SA, Taylor GK, Hernandez P, Appel RD, Binz PA, Goodlett DR. On the benefits of acquiring peptide fragment ions at high measured mass accuracy. J. Am. Soc. Mass Spectrom. 2008;19:891–901. doi: 10.1016/j.jasms.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makarov A, Denisov E, Lange O, Horning S. Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. J. Am. Soc. Mass Spectrom. 2006;17:977–982. doi: 10.1016/j.jasms.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 45.Mcluckey SA, Vanberkel GJ, Glish GL. Reactions of Dimethylamine with Multiply Charged Ions of Cytochrome-C. J. Am. Chem. Soc. 1990;112:5668–5670. [Google Scholar]
- 46.Winger BE, Lightwahl KJ, Smith RD. Gas-Phase Proton-Transfer Reactions Involving Multiply Charged Cytochrome-C Ions and Water under Thermal Conditions. J. Am. Soc. Mass. Spectrom. 1992;3:624–630. doi: 10.1016/1044-0305(92)85003-3. [DOI] [PubMed] [Google Scholar]
- 47.Qu J, Jusko WJ, Straubinger RM. Utility of cleavable isotope-coded affinity-tagged reagents for quantification of low-copy proteins induced by methylprednisolone using liquid chromatography/tandem mass spectrometry. Anal. Chem. 2006;78:4543–4552. doi: 10.1021/ac0521697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dongre AR, Jones JL, Somogyi A, Wysocki VH. Influence of peptide composition, gas-phase basicity, and chemical modification on fragmentation efficiency: Evidence for the mobile proton model. J. Am. Chem. Soc. 1996;118:8365–8374. [Google Scholar]
- 49.Diehnelt CW, Dugan NR, Peterman SM, Budde WL. Identification of microcystin toxins from a strain of Microcystis aeruginosa by liquid chromatography introduction into a hybrid linear ion trap-Fourier transform ion cyclotron resonance mass spectrometer. Anal. Chem. 2006;78:501–512. doi: 10.1021/ac051556d. [DOI] [PubMed] [Google Scholar]
- 50.Miranda TB, Miranda M, Frankel A, Clarke S. PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J. Biol. Chem. 2004;279:22902–22907. doi: 10.1074/jbc.M312904200. [DOI] [PubMed] [Google Scholar]
- 51.Lee JH, Cook JR, Yang ZH, Mirochnitchenko O, Gunderson SI, Felix AM, Herth N, Hoffmann R, Pestka S. PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J. Biol. Chem. 2005;280:3656–3664. doi: 10.1074/jbc.M405295200. [DOI] [PubMed] [Google Scholar]
- 52.Scigelova M, Makarov A. Orbitrap mass analyzer--overview and applications in proteomics. Proteomics. 2006;6(Suppl 2):16–21. doi: 10.1002/pmic.200600528. [DOI] [PubMed] [Google Scholar]