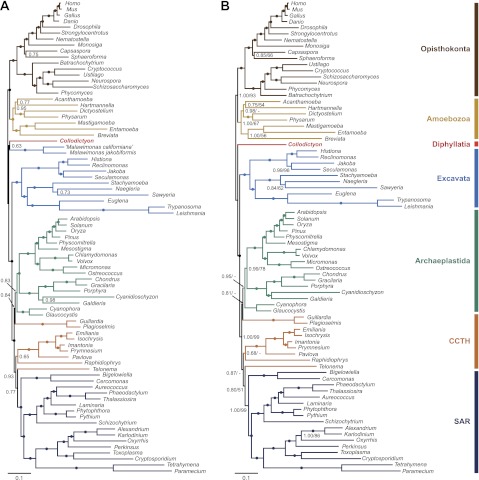
Fig. 5.
Bayesian phylogeny of Collodictyon constructed from 124 genes after removal of the fastest evolving sites. The consensus topology was calculated under the CAT model from 18,000 saved trees after discarding the first 6,000 cycles as burn-in. Branches showing 1.00 PP are marked by filled circles. The branch length of Entamoeba is shortened by 50% to save space. (A) Tree topology inferred from the trimmed alignment with the 20% fastest evolving sites removed (marked by gray rectangles in fig. 4A). Chains were considered to have converged (maxdiff = 0.104). (B) Tree topology inferred from the trimmed alignment (i.e., two Malawimonas excluded) with the 20% fastest evolving sites removed (marked by gray rectangles in fig. 4B). Chains were considered to have converged (maxdiff = 0.065). Numbers at the nodes in (B) indicate PP/bootstrap values calculated from from 100 pseudoreplicates with Phylobayes under CAT mixture model. Dashes “-” indicate bootstrap supports < 50%. CCTH is the abbreviation of Cryptophyta, Centrohelida, Telonemia, and Haptophyta. Additional statistical support values for the supergroups are shown in table 1.