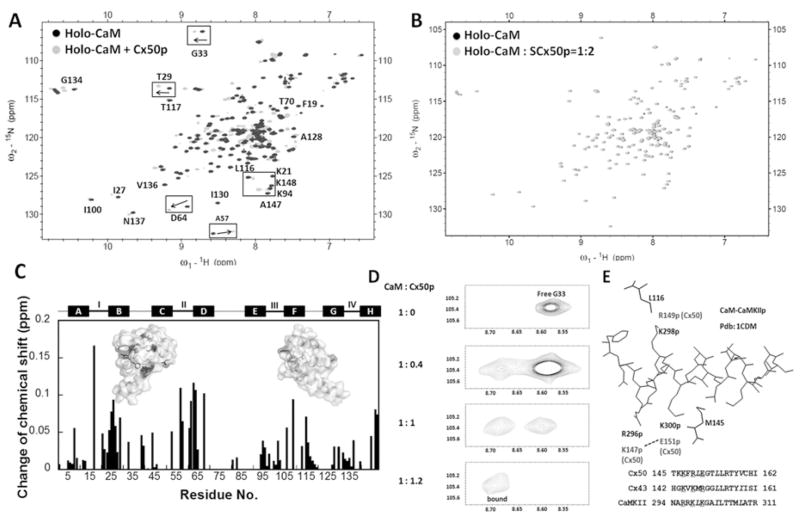
Figure 4. Monitoring the interaction between CaM and Cx50p141–166 by (1H,15N)-HSQC spectroscopy.
(A) An overlay of HSQC spectra of holo-CaM (black) with the spectrum of the holo-CaM–Cx50p141–166 (grey). (B) An overlay of the HSQC spectra of holo-CaM (black) with the spectrum of the holo-CaM–SCx50p (grey). (C) Chemical-shift perturbation in CaM induced by addition of 2-fold molar excess of Cx50p141–166. The weight-average chemical-shift change (Δδ) was calculated using eqn (5). Residues with Δδ> 0.1 p.p.m. were mapped to the three-dimensional structure of holo-CaM (PDB code 3CLN). (D) The chemical-shift change of Gly33 during titration of holo-CaM with Cx50p141–166. The disappearance of the peak (free form) was accompanied by the appearance of the corresponding peak (bound form) at a new position. (E) Structural basis of the difference in directionality of chemical shift change. The structure of the CaM–CaMKII complex (PDB code 1CDM) is shown with the peptide (inside) and the CaM residues (outside). Leu116 and Met145 of CaM are within 5 Å of Lys298p and Lys300p of the peptide respectively. These peptide positions correspond to Lys146p and Arg148p for Cx43, and Arg149p and Glu151p for Cx50, as can be seen in the superposition of their sequences. In Cx50, Glu151p may be pulled away from Met145 by Lys147p. In contrast, in Cx43, Arg148p may be pushed towards Met145 by Lys144p. These differences in peptide sequences can cause changes of chemical shifts in different directions.