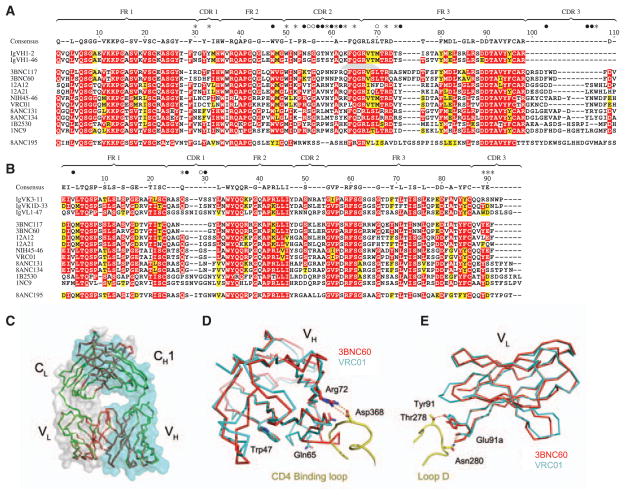
Fig. 4.
Sequence and structural conservation of HAADs. (A) Amino acid alignment of 10 selected HAADs (table S5), their germline genes, and 8ANC195. Residues are numbered according to the 3BNC60 structure. Framework (FR) and CDR regions are indicated. Red shading shows amino acid identity; yellow shows biochemical similarity. The consensus is defined by 70% similarity between the 10 selected HAADs. The consensus sequence is shown above; dashes in this sequence indicate nonconserved residues. Contact residues between VRC01 and gp120 are shown above the consensus as closed circles for main-and side-chain interactions, open circles main chain only, and stars side chains only (5). (B) As in (A) for light chains. (C, D, and E) Crystal structure of 3BNC60 Fab. (C) Superimposed Cα traces of the two Fab molecules in the 3BNC60 asymmetric unit are shown in green and red. Semitransparent surfaces are used to outline the heavy (cyan) and light (gray) chains. (D) Superimposition of the 3BNC60 VH (red, Cα trace) and VRC01 VH (cyan, Cα trace) shown with a ribbon representation of the CD4-binding loop. The salt bridge between Arg71VRC01 and Asp368gp120 is shown as dashed lines. (E) Superimposition of the 3BNC60 VL (red, Cα trace). Hydrogen bonds between VRC01 and gp120 are shown as dashed lines.