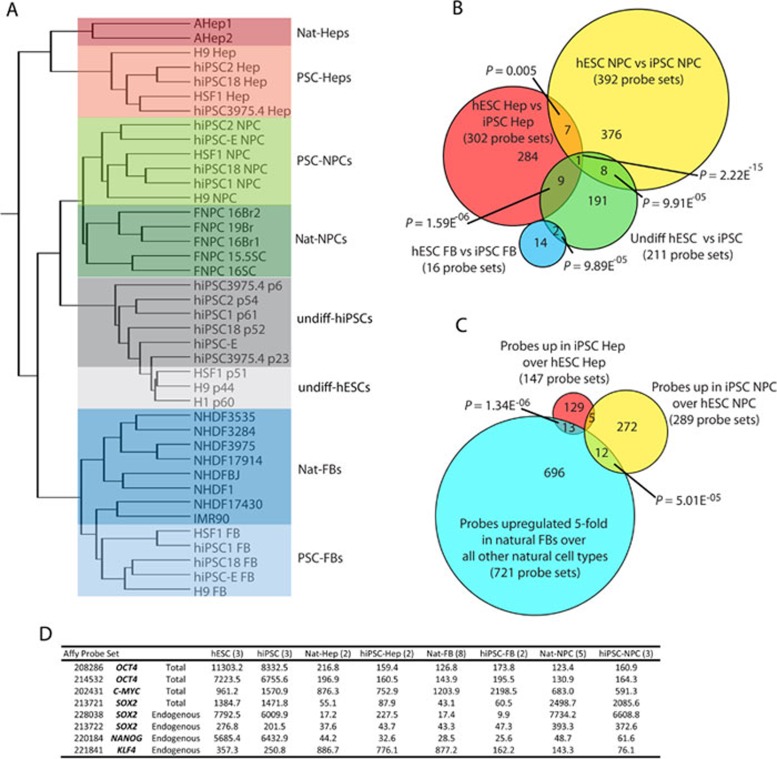
Figure 2.
Global gene expression analysis. (A) Hierarchical clustering analysis of global gene expression in undifferentiated hESCs, hiPSC, and their progeny compared to naturally derived cells. (B) Venn diagram summarizing the probe sets that were differentially expressed (t-test P < 0.01; fold change ≥1.54) between the progeny of hiPSCs versus the progeny of hESCs for each germ layer and the undifferentiated. (C) Venn diagram overlapping fibroblast signature probe sets (t-test between natural-FB and all other natural cell types; upregulated in FBs ≥5.0) with probe sets upregulated in iPSC progeny over ESC progeny for the NPC and Hep lineages. P-Values from B and C were measured by hypergeometric distribution or simulation as in 2. (D) Normalized values from microarray probe sets for the reprogramming factors used to make the hiPSCs used in this study.