Figure 1.
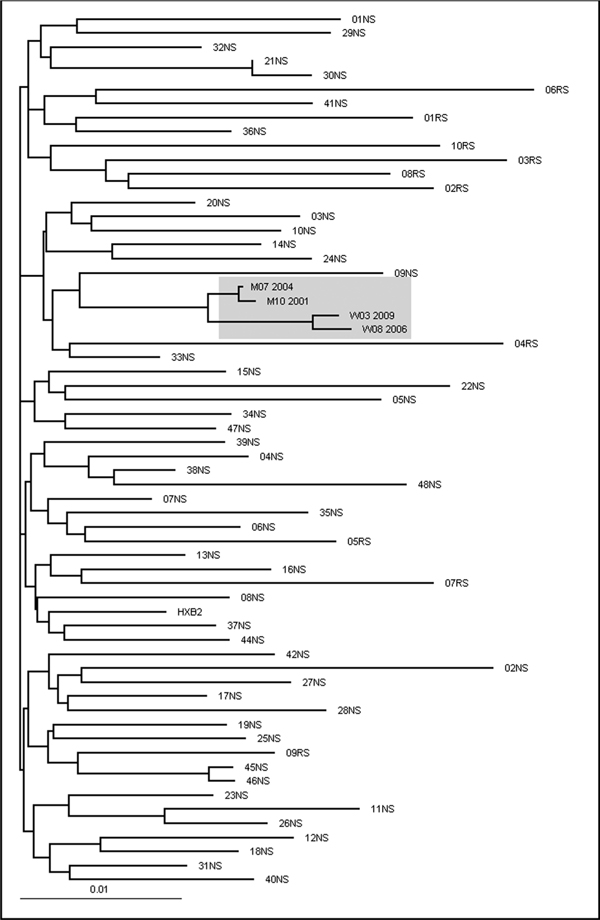
The maximum likelihood phylogenetic tree shows the close relationship between patient W and M sequences. Phylogenetic analysis was conducted on a data set of 62 HIV-1 polymerase sequences aligned through MutExt and ClustalX, including the four sequences available from patients W and M. The 48 most similar sequences to W03_2009 (01NS-48NS) and 10 per chance selected sequences (RS) from patients, obtained from the same geographic area during the same time period, were selected from a total of 6400 sequences in the Arevir database [21]. Patient W and Patient M sequences are indicated by W and M followed by the month and year of the sampling date. HXB2 is the wild type reference strain.
