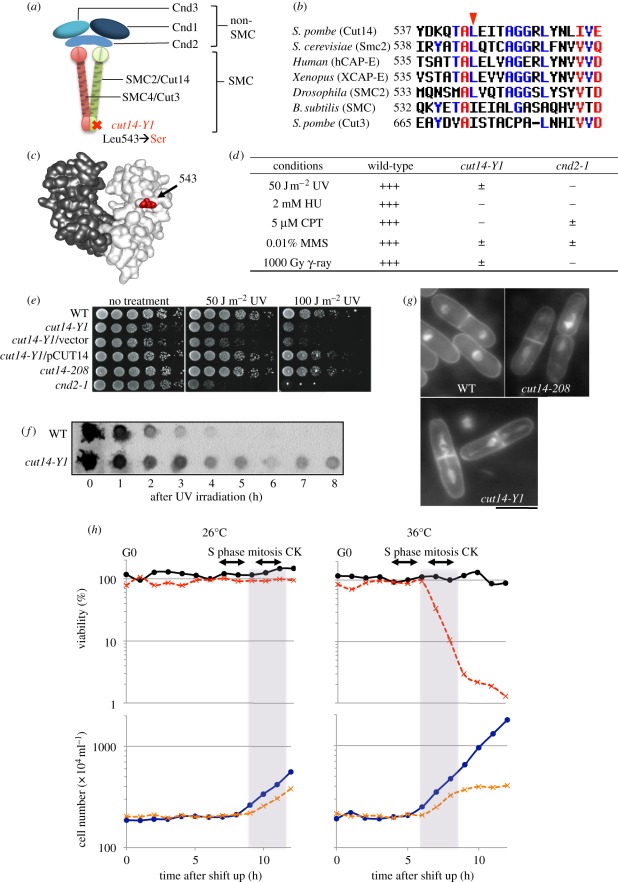
Figure 1.
Mutation site and phenotypes of condensin SMC mutant cut14-Y1. The identification of cut14-Y1: four mutant strains among 1300 ts strains examined exhibited condensation defects. Gene cloning, genetic analysis and gene sequencing established that the mutations resided in three distinct genes involved in chromosome condensation. Strain 393 was a DNA topoisomerase II top2 [26] mutant, strain 640 was a cut15 [27] (homologue of importin alpha) mutant and the remaining Y1 and 541 strains were cut14 [28,29] (SMC2 homologue) mutants. Since the cut14-Y1 strain was hypersensitive to DNA damage at 26°C (the permissive temperature), we examined whether the damage-sensitive phenotype was linked with the ts phenotype. Tetrad dissection demonstrated that the HU (hydroxyurea) and UV (ultraviolet) ray-sensitive phenotype co-segregated with the ts phenotype (electronic supplementary material, figure S1). (a) Heteropentameric condensin complex. The cut14-Y1 allele consists of a L543S substitution in the hinge. (b) Amino acid sequences of the SMC hinge that surround the mutation site (red arrowhead). (c) The mutation site (red) is shown within the three-dimensional structure of the mouse hinge domain [30]. (d) Summary of the DNA damage phenotypes of cut14-Y1 together with the previously reported response of cnd2-1 [31]. +++, normal growth; ±, very slow growth; −, no growth. (e) Wild-type (WT), cut14-Y1 and other strains were spot tested after UV irradiation at 26°C. (f) After UV irradiation (100 J m−2) at 26°C, extracts of the WT and cut14-Y1 cells harvested at intervals were immunoblotted using anti-thymine dimer antibodies. (g) The mitotic segregation defect of cut14-Y1 and cut14-208. DAPI was used to stain DNA. Scale bar, 10 µm. (h) WT and cut14-Y1 cells were first arrested at the pre-replicative G0 phase in nitrogen-deficient medium (EMM2-N) [32] at 26°C for 24 h, and then shifted to a nitrogen-replenished medium (EMM2) at 26°C (left) or at 36°C (right) for 12 h to measure cell viability (plated at 26°C) and cell number. The timing of S phase, mitosis and cytokinesis (CK) were determined by FACScan and DAPI-staining, respectively. Aliquots of the cultures were taken at 1 h intervals after replenishment, and 300 cells of each genotype were plated on three YPD plates for each time point. The plates were incubated at 26°C for 5 days, and the colony numbers were counted. Circles with solid line, WT; crosses with dashed line, cut14-Y1.