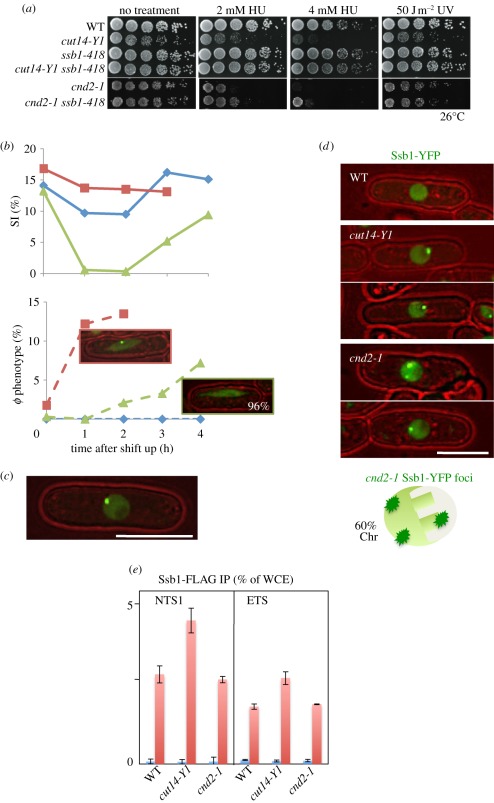
Figure 4.
The checkpoint response and nuclear localization of Ssb1-YFP differ in cnd2-1 and cut14-Y1. (a) Whereas the DNA damage sensitivities of cut14-Y1 were greatly rescued by ssb1-418 mutation, those of cnd2-1 were not. (b) WT, cut14-Y1 and cnd2-1 were cultured at 36°C for 0–4 h, and the percentage septation index (SI) and the number of cells displaying aberrant chromosome (φ phenotype) were measured. The frequency of aberrant mitotic chromosomes sharply increased in cut14-Y1 (blue diamonds with solid line, WT; red squares with solid line, cut14-Y1; green triangles with solid line, cnd2-1; top), while the appearance of such mitotic cells was delayed in cnd2-1 (blue diamonds with dashed line, WT; red squares with dashed line, cut14-Y1; green triangles with dashed line, cnd2-1; bottom). Notably, the aberrant mitotic chromosomes in cnd2-1 did not contain Ssb1 foci (96%). (c) The intense foci of Ssb1-YFP were observed in cnd2-1 cells. Note that the YFP dot (the focus) is located in the nuclear periphery chromatin region. (d) Distinct nuclear localization of the Ssb1-YFP signals in cnd2-1 and cut14-Y1. The WT cell nucleus did not show the foci of Ssb1-YFP. Two cut14-Y1 cells display the intense Ssb1-YFP foci, which are located in the nucleolar region. Two cnd2-1 cells also show the intense Ssb1-YFP foci, which are located in the non-nucleolar nuclear chromatin region. Sixty per cent of cells examined showed the nuclear chromatin localization of Ssb1-YFP foci in cnd2-1 mutant cells. (e) ChIP experiment of Ssb1-FLAG for the rDNA probe NTS1 using WT, cut14-Y1 and cnd2-1 strains. The procedures are the same employed in figure 3h. Blue bars, −antibodies; red bars, +antibodies.