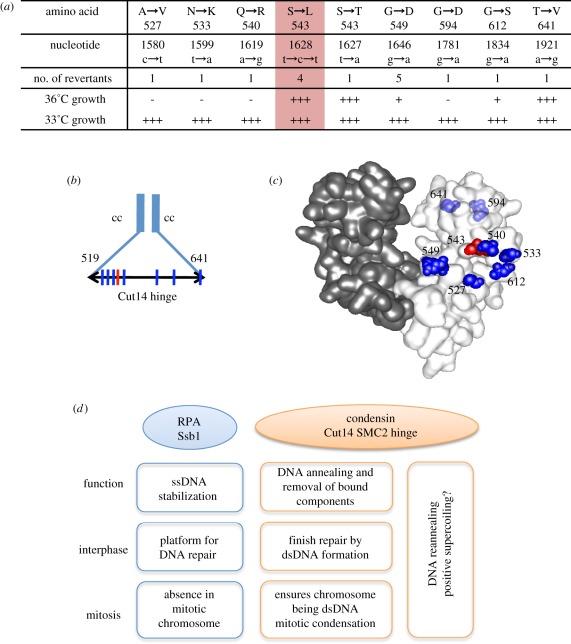
Figure 8.
The hinge of SMC2/Cut14 is a functional entity. (a) Mapping of pseudo-revertants of cut14-Y1 that formed colonies at 33°C. The four mutants are true revertants (S543L, red column) that formed the normal colonies at 36°C. See text. (b) The mutation sites are shown by the vertical lines. The hinge region is between the residues 519 and 641. cc, coiled-coil. (c) Location of the mutation sites in the three-dimensional structure of the mouse condensin hinge [30]. The amino acid residue number is adapted for the S. pombe Cut14. The original cut14-Y1 mutation site is indicated by the red colour, while the second suppressing mutation sites are shown by blue. The residues 594 and 641 situating behind are faded. (d) A diagram depicting the relationship between the condensin SMC Cut14 hinge and Ssb1. Condensin preferentially binds to ssDNA [30,34] and promotes annealing to complementary ssDNA in vitro, and appears to oppose the action of RPA. RPA acts as a platform for various proteins involved in DNA metabolism, such as damage repair and replication through ssDNA stabilization [44–46]. The role of condensin in damage repair remains unclear, but we propose that it may be required for completing/exiting repair processes by removing RPA and forming dsDNA through reannealing.