Figure 2.
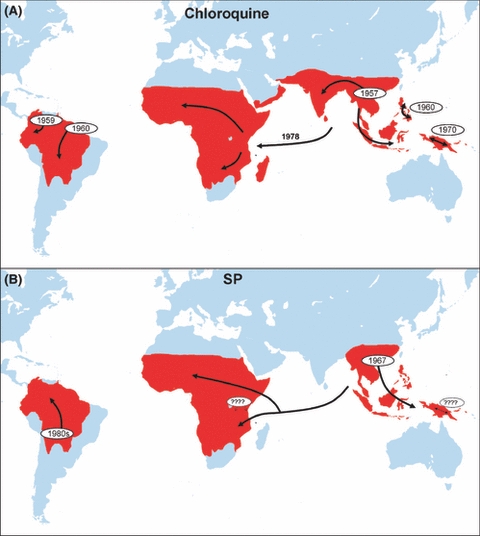
The history of chloroquine and high level pyrimethamine–sulphadoxine (SP) resistance inferred from molecular evolution studies. Chloroquine resistance has spread globally from selective sweeps from five independent origins, none of them in Africa where the health burden of drug resistance is greatest. Resistance to SP is tracked by analyses of the dhfr gene which primarily confers resistance to the pyrimethamine component. The timing of two of the independent origins is unclear. SP resistance may have several local origins in Kenya (denoted by ‘????’), but the majority of SP-resistant infections are a consequence of a selective sweep from a single origin in South East Asia. Figure 2A is redrawn from Wellems (2004), Fig. 2B is a summary of data from Cortese et al. (2002), Nair et al. (2003), Roper et al. (2003, 2004), McCollum et al. (2006, 2007, 2008), Maiga et al. (2007), Mita et al. (2007), Hayton and Su (2008), Saito-Nakano et al. (2008).
