Abstract
Although traditionally separated by different aims and methodologies, research on agricultural and evolutionary problems shares a common goal of understanding the mechanisms underlying functionally important traits. As such, research in both fields offers potential complementary and reciprocal insights. Here, we discuss adaptive stress responses (specifically to water stress) as an example of potentially fruitful research reciprocity, where agricultural research has clearly produced advances that could benefit evolutionary studies, while evolutionary studies offer approaches and insights underexplored in crop studies. We focus on research on Solanum species that include the domesticated tomato and its wild relatives. Integrated approaches to understanding ecological adaptation are particularly attractive in tomato and its wild relatives: many presumptively adaptive phenotypic differences characterize wild species, and the physiological and mechanistic basis of many relevant traits and environmental responses has already been examined in the context of cultivated tomato and some wild species. We highlight four specific instances where these reciprocal insights can be combined to better address questions that are fundamental both to agriculture and evolution.
Keywords: adaptation, agriculture, ecological genetics, gene structure and function, molecular evolution, population genetics – empirical, quantitative genetics
Introduction
Darwin (1859) pointed to the value of using domesticated plant and animals as models for understanding evolution, particularly the variability observed within and between lineages and the great power of artificial selection in effecting character change. Contemporary research in crop and other agricultural contexts continues to provide insights into understanding evolutionary processes (e.g. Ross-Ibarra et al. 2007), but evolutionary studies can likewise provide reciprocal insights into agricultural research (e.g. Alonso-Blanco et al. 2005, 2009). Here, we illustrate one example of how crop science can stimulate evolutionary research on genetics and mechanisms of adaptation in natural systems, and where evolutionary and ecological approaches might facilitate the search for agronomically valuable traits. We focus primarily on domesticated tomato and related wild species. This case study illustrates the value of contemporary interchanges between applied and evolutionary research for dissecting both specific mechanisms of trait adaptation and broad genetic and evolutionary patterns that are characteristic of adaptive change.
A primary focus of agricultural research and breeding is uncovering and exploiting agronomically valuable traits. These studies can be valuable to evolutionary biologists because they provide detailed insight into traits that are potentially fitness related under natural conditions. First, crop studies help evolutionary biologists winnow out the relevant physiological mechanisms that might contribute to adaptation in the wild. Secondly, agricultural researchers have uncovered the molecular genetic basis for some of these traits, including pathways that contribute to the normal functioning of developmental and physiological processes. This detailed physiological and molecular understanding is frequently missing in more classical ecological and evolutionary approaches to studying of adaptation (but for more recent advances see, for example, Alonso-Blanco et al. 2009; Dalziel et al. 2009). In contrast, ecological and evolutionary studies focus primarily on describing natural genetic variation within and among species and on identifying the evolutionary forces that shape this genetic variation. Because these studies are interested in organism-, population-, and species-level factors that facilitate or constrain evolutionary change, they frequently assess suites of traits and the important associations/interactions between them. Their goal is to test general theoretical predictions rather than generate an exhaustive mechanistic understanding of any one trait.
The different primary goals of evolutionary versus agricultural studies have produced different traditions of methodology and analysis. Nonetheless, these two traditions also share a common aim: understanding the mechanisms underlying functionally important traits. Within this general aim, several important research questions are also shared including, for example, questions of how traits are optimized and/or constrained, and how they are influenced by resource and/or physiological trade-offs to maximize yield (in the case of agriculture) or fitness (in the case of evolution) (e.g. Chapin et al. 1993). As such, while the specific goals of each research program differ, these parallel sub-fields offer evident reciprocal value.
To illustrate this potentially fruitful research reciprocity between agricultural and evolutionary studies, we focus here on traits important for adaptation to abiotic stress, especially drought stress. Other papers have explored research synergies at the intersection of evolutionary and breeding/functional studies for other traits, including developmental processes such as flowering time and plant architecture (e.g. Alonso-Blanco et al. 2009), however drought responses have been comparatively underexplored (although see Nevo and Chen 2010). Nonetheless, a substantial component of modern crop research is focused on the identification and exploitation of genetic traits that improve yield under marginal abiotic conditions, including drought (e.g. Passioura 1996; Richards 1996; Araus et al. 2002; Chaves and Oliveira 2004). Conversely, among evolutionary geneticists there is intense interest in the genetic basis and evolutionary history of traits that allow the exploitation of ecologically ‘difficult’ habitats, including those with limited or ephemeral water, extended periods of climatic (heat or cold) extremes, and soil toxicity. Abiotic climatic factors like water availability and temperature are fundamental determinants of plant distribution and abundance (Boyer 1982) and act as strong selective forces on the evolution of plant physiology and form (Ehleringer and Monson 1993; Bohnert et al. 1995; Bray 1997). Therefore, understanding adaptive responses to water deficit might be a particularly fruitful area of research reciprocity between agriculture and evolutionary biology. Throughout, we focus primarily on the plant group that contains the domesticated tomato, Solanum Section Lycopersicon, while referencing other model systems when appropriate.
Tomato as a system to understand genetics and adaptation to abiotic stress
In many ways, tomato and its wild relatives (Solanum Section Lycopersicon) exemplify the reciprocal insights that parallel agricultural and evolutionary studies could offer into understanding adaptive abiotic responses (Moyle 2008). The group forms a relatively small monophyletic clade within the large and diverse Solanaceae family (D'Arcy 1979), consisting of 14 closely related species or subspecies including the domesticated tomato, Solanum lycopersicum (formerly L. esculentum) (Peralta et al. 2008). Domesticated tomato is an economically important crop that is grown throughout the world, in countries that range geographically from the tropics to far northern and southern latitudes. Because of the geographical breadth of its cultivation (and the expense of maintaining artificial fertilization, pesticide, and watering regimes), there is intense interest in uncovering and exploiting genetic traits that contribute to increased yield under a large range of environmental conditions, including temperature and water availability extremes, and across variation for natural biotic pests (Zamir 2001; Fernie et al. 2006; Bai and Lindhout 2007; Barone et al. 2009). In comparison to domesticated tomato, wild tomato species span a vast range of climatic, biogeographic, and environmental variation, from temperate deserts to wet tropical rainforests (Peralta et al. 2008). The natural range of wild tomatoes is Ecuador to Chile, along the eastern and western Andean slopes, and coastal region of northwest South America. One species is also found in central America, and one is endemic to the Galapagos Islands. Due to a recent dynamic geological history, this region encompasses dramatic environmental gradients (e.g. sea-level to >3000 m elevation, 50 mm to >4000 mm annual precipitation) and geological and geographical structure (Young et al. 2002). Despite recent divergence of the tomato group (between ∼2.7 and <7 million years; Nesbitt and Tanksley 2002; Kamenetzky et al. 2010), each species appears to display a characteristic geographical distribution pattern and habitat preference across this environmentally diverse region (Rick 1973, 1978, 1979; Nakazato et al. 2010). Species are also morphologically diverse, and some traits are likely adaptive responses to local habitats (e.g. Rick 1973, 1976; Patterson et al. 1978; Rick 1978; Vallejos 1979; Bloom et al. 2004; Nakazato et al. 2008, and see below), suggesting that abiotic ecological conditions likely played an important role in phenotypic evolution and speciation in this group.
In addition to this natural biological diversity in relevant traits, Solanum Section Lycopersicon also has logistical advantages as a model system for understanding the genetic basis of fitness-related traits. All members of the clade are closely related diploids (2n = 24) (Peralta and Spooner 2001; Nesbitt and Tanksley 2002) that share a high degree of synteny (Quiros 1991; Chetelat and Ji 2007), and are to some degree intercrossable (Rick 1979); therefore species differences are amenable to genetic analyses. The group is also the subject of extensive genomic development due to the agronomic importance of tomato and its close relationship with other Solanum species including potato, capsicum, and eggplant. Current genomic tools include a high density genetic linkage map in S. lycopersicum, multiple permanent quantitative trait locus (QTL) mapping populations, an ongoing genome sequencing initiative, BAC libraries, an EST database, microarray platforms, and thousands of molecular markers appropriate for use in domesticated and wild species (Frary et al. 2005; Mueller et al. 2005; Foolad 2007; Barone et al. 2009; SOL Genomics Network: http://solgenomics.net/). As such, tomato has both the requisite genetic tools and ecological diversity to address the genetics of drought responses, both from plant breeding and evolutionary perspectives (see also Moyle 2008).
Drought stress responses in crop breeding and genetics
Studies in many agricultural systems indicate that drought adaptation is complex (Collins et al. 2008) and can involve alternative strategies including dehydration tolerance, dehydration avoidance, and drought escape (Ludlow 1989). In addition, traits relevant to drought responses can be highly environmentally plastic (e.g. stomatal density is highly plastic to light environment; Gay and Hurd 1975; Ticha 1982), complicating efforts to undercover their genetic basis. Nonetheless, these studies have been successful in identifying specific traits and genetic pathways that can contribute to drought adaptation in crop species. For example, mechanisms of drought tolerance are known to include leaf traits, root traits, and osmotic regulatory responses (e.g. Richards 1996; Bray 1997; Chaves and Oliveira 2004; Collins et al. 2008; Nevo and Chen 2010), as well as elevated water-use efficiency (WUE) measurable as either instantaneous or lifetime WUE (Condon et al. 2004).
Here, we highlight two areas in which tomato crop research has offered valuable insights into understanding mechanisms and genetics of drought responses: uncovering physiological mechanisms, and identifying contributing genetic pathways.
Physiological mechanisms
In tomato, most ecophysiological analyses have focused on genetically based variation within domesticated tomato (including between mutant types) (Hsiao 1973). Studies of drought within domesticated tomato have been primarily concerned with the influence of soil water deficits on productivity and yield (e.g. Wudiri and Henderson 1985; Mitchell et al. 1991; Renquist and Reid 2001; Yoo et al. 2009) but also reveal a range of physiological mechanisms that underlie tomato drought responses, including osmotic and developmental responses to water deficit (e.g. Rudich et al. 1977; Stevens and Rudich 1978; Thakur 1990; Tan 1993; Reid and Renquist 1997; Romero-Aranda et al. 2001) that are mostly consistent with dehydration avoidance responses. Some physiological studies have also extended outside domesticated varieties, to evaluate wild germplasm for traits that could contribute to yield under water-limited conditions (e.g. Kahn et al. 1993; Martin et al. 1999; Maldonado et al. 2003; Bloom et al. 2004). For example, Solanum pennellii has cactus-like features including succulent leaves and very shallow spreading roots (Rick 1973). Solanum chilense develops extremely deep roots, possibly to absorb water stored in deep soil from occasional flooding (Rick 1973). Wild species of tomato also differ in drought-related traits such as specific leaf area (SLA, a measure of leaf thickness associated with propensity for transpirational water loss) (e.g. Easlon and Richards 2009).
In terms of physiological drought mechanisms, perhaps the best understood wild tomato species is S. pennellii. Solanum pennellii has greater WUE (defined as the ratio of carbon assimilated to water lost) than domestic tomato (Martin and Thorstenson 1988; Kebede et al. 1994; Martin et al. 1999) and other self-compatible wild species including Solanum pimpinellifolium (Easlon and Richards 2009). Increased WUE and ability to withstand drought is primarily due to leaf anatomy (smaller area and lower stomatal density) and stomatal behavior that conserves water (Kebede et al. 1994). Data conflicts as to whether S. pennellii has significantly lower SLA (Kebede et al. 1994; Torrecillas et al. 1995; Comstock et al. 2005; P. Davis, pers. comm.), but it is certain that they have smaller, more succulent leaves (Kebede et al. 1994). Accordingly, agricultural studies provide important insights into the physiological mechanisms that likely contribute to natural adaptive responses to water deficit in this species. Conversely, drought escape, osmotic adjustment or increased cell elasticity (Torrecillas et al. 1995), rapid induction of drought response (Kahn et al. 1993), and extensive root systems (Martin et al. 1999) have not been found to contribute to drought adaptation in S. pennellii. In terms of understanding the molecular and genetic mechanisms of trait variation, the exclusion of these potential physiological mechanisms is equally useful in ultimately identifying the underlying basis of drought adaptation in this species.
Candidate genes
Basic scientific and agronomic interest in developing drought tolerant crops has uncovered numerous genes affecting plant–water relations that are potential candidates for adaptive differentiation in nature (e.g. Bartels and Sunkar 2005). The absiscic acid (ABA) biosynthesis pathway is of particular interest in tomato. ABA is a hormone synthesized primarily in roots and transported to leaves during water-deficit stress (Thompson et al. 2007a; Cutler 2009). It plays a major role in drought response, especially stomatal closure. The level of ABA in the plant is finely tuned by the relative rates of biosynthesis and catabolism (Nambara and Marion-Poll 2005). Thus, adaptive differences in stomatal behavior evident between tomato species (see above) could reasonably be caused by changes in metabolic enzyme activity. The ABA metabolic pathway is also small (six genes) and has been studied in domestic tomato more than any species excepting Arabidopsis. Previous work has demonstrated that the enzyme NCED is the rate-limiting step in ABA biosynthesis in tomato (Thompson et al. 2000, 2004). Furthermore, experimentally increased expression of NCED led to increased drought tolerance under greenhouse conditions (Thompson et al. 2007b). Given these observations, ABA metabolic enzymes are especially promising candidate genes to investigate in tomato.
Evolutionary studies of abiotic variation in wild tomato species
Evolutionary studies of adaptation in plants have traditionally been focused on demonstrating the role and efficacy of natural selection in producing environmentally adapted genotypes (e.g. Stebbins 1952). For example, classical reciprocal transplants and/or common garden experiments reveal interspecific variation in tolerance to water stress that is associated with historical environmental water availability, although few of these studies have investigated the underlying physiological mechanisms responsible (e.g. Dudley 1996a,b; Heschel et al. 2002). More recently, studies have begun to address the genetic basis of natural variation in drought-stress responses, in terms of the number, individual effect, and genomic location of loci contributing to drought-related traits (McKay et al. 2008 and references therein). Here, we highlight two areas in which ‘evolutionary’ approaches and/or tools can add insight to understanding natural mechanisms of response to water availability in tomato.
Quantifying natural environmental and trait variation for drought responses in wild species
Geographical differences in water availability are expected to select for genetic differences in drought tolerance among population and species in the wild (Stebbins 1952). One challenge for understanding natural adaptation to environmental variation is obtaining quantitative climatic data on the native environments of each group of interest. Among wild tomato species, we have used Geographical Information Systems (GIS) to quantify the mean and range of environmental conditions associated with the geographical distribution of each species (Nakazato et al. 2010). These analyses integrate location data from geo-referenced occurrence records with global climate databases, and can be used to identify climatic differences between groups of interest (Kozak et al. 2008). Among tomato species, we find that each has a unique combination of characteristic environmental variables (Nakazato et al. 2010; Fig. 1); moreover, the most prominent axis of species differentiation is habitat aridity (e.g. mean annual rainfall) (Nakazato et al. 2010). This indicates that response to water availability likely influences the distribution and evolution of almost all wild tomato species, and implies that species are likely to be differentiated with respect to drought response traits.
Figure 1.
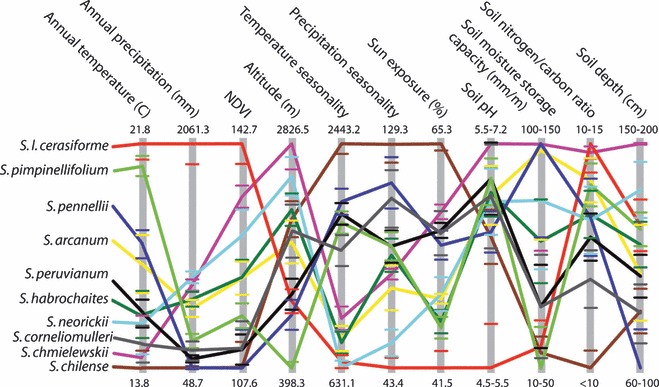
Environmental variation among tomato species determined from Geographical Information Systems analysis of bioclimate data (modified from Nakazato et al. 2010). Different colors represent different species; vertical gray bars represent 10 environmental variables plus altitude, with population mean values for each species indicated where each species line crosses the vertical bar. Numbers indicate the maximum and minimum of the mean species values for each environmental variable. Short horizontal lines on the gray bars show the 95% confidence intervals of the environmental values for species with respective colors (lines above and below the maximum and minimum values are omitted). NDVI is Normalized Difference Vegetation Index, and is an estimate of vegetation cover based on land surface reflectance spectra, as measured with satellite imagery.
The application of these ‘macroecological’ (Brown 1999) approaches extends beyond quantifying environmental variation within and between species, and identifying ecological factors most important for differentiation. In particular, measurable variation in native climatic conditions can be used to predict patterns of natural adaptive trait variation within and between species – hypotheses that are testable with direct analyses of natural quantitative genetic variation. For example, we assessed whether morphological and physiological trait variation within and between two wild tomato species (assessed under common garden and drought manipulation conditions) matched climatic variation identified with GIS analyses (Nakazato et al. 2008). We confirmed that several eco-physiological traits show significant trait–climate associations among climate-differentiated populations of S. pimpinellifolium, including a strong association between native precipitation and whole-plant tolerance to water deficit (Nakazato et al. 2008; Fig. 2). Conversely, we also identified traits that were not associated with gradients in climatic factors or with environmental manipulations in these studies. These analyses can therefore be useful for both identifying the specific environmental factors (e.g. water availability) likely responsible for natural adaptive diversification and narrowing the possible underlying phenotypic mechanisms (candidate traits) involved.
Figure 2.
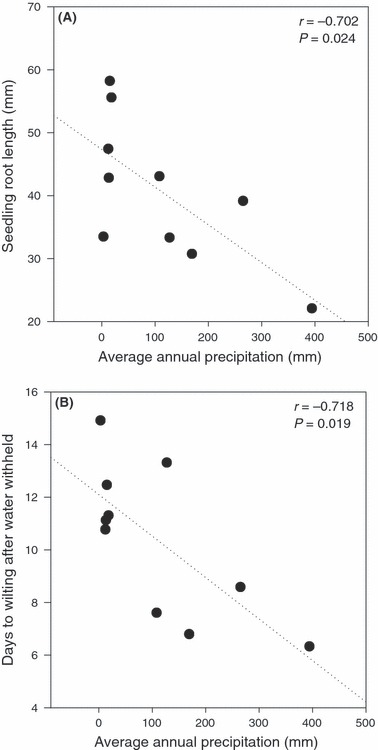
Trait–climate associations between quantitative genetic traits and bioclimate variables generated using Geographical Information Systems (modified from Nakazato et al. 2008). As described in Nakazato et al. (2008), 10 populations of Solanum pimpinellifolium were assessed for quantitative genetic variation in a common garden environment and under drought manipulation. Top panel (A) shows the relationship between native mean annual precipitation (mm) experienced by each population, and the average seedling root length when germinated under common garden conditions. Bottom panel (B) shows the relationship between native annual precipitation and the number of days to wilting in juvenile (prereproductive) plants from each population, once water was withheld. Both significant relationships suggest that traits associated with drought responses are preferentially found in populations with low native rainfall, consistent with natural selection for these population differences.
Apart from studies of natural genetic variation, the use of these GIS approaches to identify species climatic envelopes and environmental tolerances could also have more direct applications of interest to crop scientists, for example the quantitative prediction and identification of wild germplasm likely to carry traits of agricultural value (see below). Natural ecological and evolutionary systems are a source for a vastly expanded range of trait variation; this variation can be valuable in mechanistic analyses, including for uncovering the underlying genetic and molecular mechanisms responsible for agriculturally valuable quantitative trait variation (e.g. Alonso-Blanco et al. 2005, 2009; Johal et al. 2008; and see below).
Identifying candidate loci using tests of molecular evolution
Reverse genetics approaches (e.g. knockouts, knockdowns, targeted gene replacement) are the gold standard for assessing and confirming the functional role of a gene (e.g. Fernandez et al. 2009). Because of the technical efforts involved, these approaches have traditionally relied on strong experimental evidence to identify likely candidate loci a priori. Evolutionary approaches offer alternative methods for identifying potential candidates for important physiological responses, by using patterns of molecular genetic variation to identify DNA sequences with unusual patterns of evolutionary change, and/or to reveal or exclude the action of natural selection on specific loci (e.g. Nielsen 2005; Storz 2005). These approaches are often most valuable when used to cross-evaluate loci that have been functionally identified in one model system (e.g. Arabidopsis) for their potential involvement in equivalent functional transitions or roles in a new species system (e.g. tomato), or when functional analysis in a single genotype implies that a gene might be involved in functional differentiation among multiple closely related genotypes.
Owing to an incomplete genome sequence, tests of molecular evolution have been relatively limited in tomato, although they include loci involved in pathogen resistance (e.g. Caicedo and Schaal 2004; Rose et al. 2007). In terms of loci implicated in drought responses, Frankel et al. (2003) analyzed patterns of molecular genetic differentiation among six wild tomato species in ASR2, a locus known to be involved in drought and temperature stress responses in domesticated tomato. They detected evidence for non-neutral evolution (excess nonsynonymous changes) along branches leading to two wild taxa (S. chilense and S. arcanum), and suggested that these patterns might be due to selection for drought adaptation. In a follow-up analysis of intraspecific molecular genetic variation, Giombini et al. (2008) detected evidence for non-neutral evolution within these two wild species, and speculated that differences in nucleotide diversity between these species might be due to different selective regimes and/or demographic history.
Other studies have also implicated loci in adaptive environmental differences between tomato species, thereby identifying potential candidates for both agricultural and evolutionary studies. For example, comparing homologous sequences generated from EST libraries of three tomato species [S. lycopersicum (tomato), S. pennellii, and S. habrochaites], Jimenez-Gomez and Maloof (2009) found evidence for positive directional selection among species on 10 loci; these loci were enriched for functions related to abiotic and biotic stimulus, as defined by gene ontology categories based on Arabidopsis homologs.
Combining agricultural and evolutionary insights
Functional studies from agricultural research offer essential insights into the specific mechanisms that can underlie important trait variation. With respect to drought adaptation, functional and physiological analyses in the crop literature already provide considerable information on the morphological and physiological traits that might be of greatest adaptive importance among tomato species, as well as candidate loci for analyses of drought adaptation. Conversely, evolutionary approaches can identify general patterns of trait variation and response, and can vastly expand the repertoire of natural genetic variation with which to examine (and potentially exploit) functionally important traits. Nonetheless, these reciprocal resources have been underutilized by ecological/evolutionary geneticists and crop scientists alike. Here we highlight four areas where advances could be made towards reciprocal understanding within agricultural and evolutionary fields, with respect to drought responses in tomato.
Identifying and exploiting natural variation for crop improvement
Evolutionary analyses of natural trait variation in wild tomatoes promises to identify exploitable genetic variation that might be valuable for future crop improvement, either directly via targeted trait introgression from wild species (Zamir 2001), or indirectly via expanding the range of genetic variation that is available to understand molecular mechanisms of trait variation (e.g. Alonso-Blanco et al. 2009).
The process of domestication and subsequent artificial selection is known to have several genome-wide genetic consequences including the reduction of variation at both trait and DNA sequence levels (e.g. Bai and Lindhout 2007; Van Deynze et al. 2007). Because of the close relationship among all species in the clade, and the vastly reduced genetic variation within the cultivated tomato, wild tomato species have been the source of multiple agriculturally valuable traits for crop improvement, such as disease resistance and vitamin content in fruits (Zamir 2001; Gur and Zamir 2004; Bai and Lindhout 2007; Lippman et al. 2007). Tomato breeders have previously used climatic or environmental factors to guide the choice of wild germplasm to evaluate for crop improvement (Atherton and Rudich, 1986), however GIS-based environmental characterization (see above) offers a comprehensive quantitative approach to identifying populations most likely to exhibit adaptive traits of interest. These approaches can also simultaneously take into account multiple environmental variables, such as native water availability, temperature, and soil traits, thereby potentially minimizing (or maximizing) the influence of traits that affect more than one environmental factor. As such, in close relatives of economically important species, these approaches can provide a more quantitative and precise set of predictions about which sources of wild germplasm might provide the most useful natural genetic variation.
This trait variation is likely to exceed that found in domesticated varieties both quantitatively and qualitatively. In tomato, investigations of natural variation already show that different wild species likely have alternative mechanisms of adaptation to the same abiotic conditions. For example, physiological water stress responses differ between S. pennellii and S. chilense (see above), making both species sources of new and unique traits for possible crop improvement. In addition to simply identifying traits of agronomic interest, this natural genetic variation can also provide the raw material for new insights into the diversity of molecular mechanisms that determine important trait variation. In other crop and model systems (e.g. Alonso-Blanco et al. 2009), the analysis of genetic variation has provided unprecedented information on the molecular mechanisms that determine important adaptive responses, insights that are unavailable when relying solely on domesticated varieties or mutants for experimental material. For example, Nevo and Chen (2010 and references therein) review studies identifying traits related to drought and salinity stress from multiple wild cereal species.
Finally, GIS-based information can also be combined with species distribution modeling (SDM) to provide geographical predictions of species habitat suitability (see Nakazato et al. 2010). These predictions identify geographical locations that are likely (or unlikely) to be suitable habitats for a species of interest (Kozak et al. 2008). These predictions can be used as geographic guides for future collecting efforts for new, potentially valuable wild germplasm. In this way, GIS environmental characterization can provide quantitative tools to evaluate useful germplasm in existing collections, and SDM can provide an assessment of promising areas for future germplasm collections.
Dissecting the genetic basis of adaptation
Evolutionary biologists are still in the incipient stages of uncovering the genetic basis of adaptation in wild organisms. For some exemplar systems, such as pelvic reduction in threespine stickleback fish (e.g. Chan et al. 2010) or flower color (Rausher 2008), the molecular genetic basis of adaptation is partially known. Progress in these systems owes much to the fact that they focus on ecologically relevant phenotypes that have a relatively simple (e.g. loss-of-function) genetic basis. In contrast, the genetic basis of adaptation for complex traits composed of many interacting constituent parts is in its infancy, for technical as well as biological reasons. Trivially, uncovering the genetic basis of complex traits may be practically difficult because the pertinent phenotypes are difficult to measure. Other complications, such as pleiotropy, linkage, and sensitivity to genetic (epistasis) and external (plasticity and G × E) environments, are more biologically meaningful, but nevertheless obscure inference about causal connections between loci, traits, and adaptation. Importantly, it is this uncharted empirical landscape that has been the purview for most theory of adaptation (Fisher 1930; Orr 2005). Bridging the gulf between theory and data will require methods to address complex, ecologically important traits.
Anatomizing complex traits into multiple, easily measured phenotypes, each of which may have a relatively tractable genetic architecture, is a promising approach. However, it may be difficult to know which phenotypes are important, as there are often multiple suites of traits that are functionally similar. In this context, agricultural research can provide valuable information on which traits are likely candidates for adaptive differentiation in nature. For instance, crop research on physiological dehydration avoidance in tomato species allows approaches like QTL mapping to be more precisely targeted to those traits most likely to reflect adaptive differentiation between species. For example, despite clear differences in tolerance between S. pennellii and S. lycopersicum (see above), functional traits enhancing performance during drought have been mapped to a very limited degree among these and other tomato species (Martin et al. 1989; Foolad et al. 2003; Xu et al. 2008). However, using currently available genetic tools, such as recombinant mapping population, it is relatively straightforward to identify QTL that underpin these physiological and functional differences (e.g. Muir and Moyle 2009). These more targeted efforts, informed by physiological studies in agriculture but incorporating wild species variation, make dissecting the genetic basis of drought adaptation highly promising in tomato and similar systems. For example, for numerous species of cultivated cereal grains, there are many QTL mapping studies of drought tolerance traits that capitalize on natural variation (see Chen et al. 2010 and references therein for a current account).
The candidate trait approach also enables targeted examination of interactions between traits (pleiotropy) and between genes (epistasis) based on a priori theoretical and/or mechanistic expectations. For example, for plant drought responses, there is little reason to expect a strong mechanistic connection between nontranspirational seedling growth and adult WUE. Conversely, leaf size and stomatal density are usually negatively correlated within species, as developmental patterning is determined prior to leaf expansion. Understanding this mechanistic connection suggests explicit expectations for constraints on adaptive response. For example, dehydration avoidance entails both reduced transpirational area and decreased stomatal density (as observed in differences between S. pennellii and S. lycopersicum), implying that multiple mutations should be necessary for selection to break this negative trait correlation. Similar expectations can be generated with respect to epistasis. For example, theory predicts that genes in the same pathway, and therefore affecting the same trait, may exhibit antagonistic epistasis (Phillips 2008), leading to an expected excess of such interactions between QTL underlying the same trait, but not between QTL for different traits. Interestingly, in a QTL analysis of ecophysiological traits differentiating S. pennellii and S. lycopersicum (Muir and Moyle 2009), we detected an abundance of antagonistic epistatic interactions between loci influencing SLA and whole-plant drought responses, consistent with these theoretical predictions.
Together, these complex trait and locus interactions will determine the trajectory of and constraints on adaptive evolution in the wild. Assessing their relative importance relies on experimental systems, like tomato, in which mechanistic and functional data can be combined with predictions from evolutionary genetic theory. Of course, the resulting data will not only offer insights into the genetics of adaptation in the wild. In an interesting inversion of Darwin's original analogous reasoning between natural and artificial selection, understanding how genetic architecture constrains and facilitates of natural selection can equally act as a guide for informing expectations about the effects of artificial selection on trait variation subject to pleiotropy, epistasis, and constraint.
Assessing functional and evolutionary trade-offs
Integrative approaches to studying trait variation in tomato can also be used to address long-standing questions in the ecophysiological and evolutionary literature (e.g. Chapin et al. 1993). For example, trade-offs arising from biophysical constraints are thought to prevent simultaneous adaptation to multiple contrasting environments (Lambers et al. 2008). Drought adaptation is a canonical example because water loss occurs during photosynthesis, ‘constitut[ing] a fundamental trade-off, evidenced by a negative correlation, between fixing carbon and saving water’ (Arntz and Delph 2001). However, evidence that trade-offs play an important role in divergence and coexistence between closely related species or populations is equivocal (Ackerly et al. 2000; Geber and Griffen 2003; Hereford 2009). Genetic studies of drought adaptation also give conflicting results. McKay et al. (2003) found a trade-off (antagonistic pleiotropy) between two drought escape traits: WUE and flowering time. Conversely, adaptation to drought in the wild oat Avena barbata appears to entail no fitness cost in mesic environments (Latta 2009).
An ecophysiological trade-off is evidenced by a negative cross-environment correlation related to the value of a functional trait. For example, if smaller leaves improve growth during drought by reducing water loss, but decrease growth in well-watered conditions by limiting photosynthesis, then one could conclude there is a trade-off involved in changing leaf size. Two criteria are required for inferring a trade-off:
a negative correlation between performance in contrasting (e.g. dry and wet) environments.
provided criterion 1 is met, performance must be correlated with trait values in the expected direction to differentiate trade-offs from spurious genetic differences.
Both of these can be functionally evaluated in tomato using natural variation for drought adaptive traits. For example, seed germination success and seedling growth vary predictably in response to low and high osmotic stress, depending upon the native water conditions experienced by different species. Using germination media in which the osmotic potential can be artificially manipulated, we found that a species from more water-limited natural environments (S. pennellii), continues to show substantial germination under high osmotic stress (Fig. 3). In comparison, species that occur under more abundant water conditions [e.g. S. lycopersicum (tomato) and S. pimpinellifolium] show precipitous declines in germination rates under high osmotic stress (Fig. 3). Interestingly, in low osmotic stress conditions, seedling growth rates are retarded in the ‘drought-adapted’S. pennellii, in comparison to the other two species (data not shown). Together these results are indicative of a negative correlation between seed germination and seedling establishment in low and high osmotic stress conditions, a necessary condition (criterion 1, above) for demonstrating a functional trade-off between adaptation to alternative resource environments. This apparent trade-off might be related to species differences in seed size. The smaller seed of the drought-adapted species gives it a greater surface area to volume ratio, increasing imbibition, but decreasing maternal provisions that can be marshaled during early growth. This and alternative hypotheses can be tested with further experimental analysis of the loci underlying such traits. We are also testing for dehydration avoidance traits in S. pennellii (see above), which are likewise predicted to exhibit trade-offs. The availability of genetic tools, including recombinant lines that allow the identification of QTL, are particularly valuable in this respect.
Figure 3.
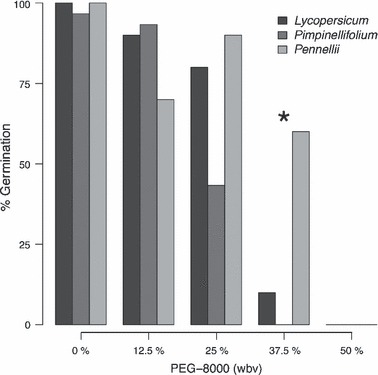
Seed germination success of different tomato species under low and high osmotic stress. Seeds were germinated on growth media with variable concentrations of PEG-8000 (polyethylene glycol-8000), a neutral polymer that replaces water in the growth media but is unable to be absorbed by plant roots. Osmotic stress increases from left to right with increasing PEG-8000 concentration. Asterisk indicates a significant difference between species (P < 0.05). Result indicate that, at high osmotic stress, dry adapted Solanum pennellii continues to show appreciable levels seed germination, in comparison to S. lycopersicum (tomato) and S. pimpinellifolium.
The classic evolutionary problem of how species cope with constraints and trade-offs reinforces what crop scientists already know about complex traits. Breeding for tolerance to a single stress may decrease yield in benign conditions or under a different stress. The proper balance of stress tolerance, yield, and other traits thus needs to be tuned to specific environmental conditions (Nevo and Chen 2010). Evolutionary studies can reveal new prospects and challenges for understanding these relationships. Over eons, natural selection has struck upon many innovative solutions to work around constraints, providing insights that may not have been gained from first principles. Conversely, consistent and evolutionarily widespread lack of trait variation in some directions may reveal unforeseen and unavoidable constraints.
Identifying repeated patterns and common functional pathways in adaptive transitions
In the future, integrating broad clade-level comparative analyses of abiotic niche evolution with a more mechanistic understanding of the genetic basis of adaptive species differences (e.g. QTL mapping and functional studies) promises to provide substantial insights into the ‘genetic architecture’ of adaptation. It is in this regard that agricultural studies have the greatest potential to contribute conceptually to evolutionary analysis – that is, via the detailed information they provide on the specific genetic mechanisms responsible for phenotypically important trait changes. Given access to this rich mechanistic information, and the ability to extend it into natural evolutionary settings, comparative evolutionary studies can identify general patterns and processes of adaptation (Harvey and Pagel 1991). These in turn can be used to address ongoing conceptual debates within evolutionary biology including, for example, whether adaptive change is enriched for specific kinds of molecular mutation (e.g. regulatory versus structural changes; Hoekstra and Coyne 2007), whether adaptation frequently involves diversification of gene duplicates (e.g. Oakley 2007; Hahn 2009), and the relative importance of G × E for natural trait expression and responses to selection (e.g. Weinig and Schmitt 2004; Sultan 2007).
For example, Kopp (2009) has advocated a meta-model approach for evolutionary genetics that takes advantage of parallel trait changes in a clade to ask whether and why convergent phenotypes are underpinned by similar genetic changes. Repeated adaptation to novel niches is a pervasive form of parallel evolution, and abiotic adaptive diversification is one instance in which multiple evolutionary transitions can be used to identify patterns in either the traits and/or underlying genetic basis of adaptive phenotypic change. For example, in tomato, patterns of molecular evolution in specific pathways contributing to drought adaptation, e.g. the ABA biosynthesis pathway, can be examined to determine the repeatability of specific evolutionary changes in both domesticated and wild lineages.
These analyses can also contribute to other complementary questions about the nature and predictability of genetic changes that underpin adaptation. For example, there is a growing literature on pathway and network evolution (Flowers et al. 2007, 2009; Rausher 2008; Alvarez-Ponce et al. 2009; Ramsay et al. 2009). Based on metabolic control theory (Kacser and Burns 1973) and similar modeling approaches, it is predicted that genetic changes affecting the function of most enzymes will not affect the phenotype, making them effectively invisible to selection. Rather, control may lie in mutations at a few key loci that, because of their position in a pathway (e.g. branch points [Eanes 1999]), can affect fitness. Within tomato, these theoretical considerations can be used to generate and test predictions about where functional genetic changes are most likely to take place within specific networks and pathways, such as the ABA biosynthesis pathway (see above). In addition to better understanding processes of natural adaptation, these analyses can also be useful for identifying which loci are more likely to be fruitful in manipulating such traits to maximize yield outcomes in agricultural settings.
Some of these conceptual questions lie directly at the intersection of evolutionary and agricultural studies (Hancock 2005; Ross-Ibarra et al. 2007; Purugganan and Fuller 2009). For example, the strength and mode of selection on individual loci during adaptation is debated, with implications for the type (regulatory versus coding) and number of mutations involved. One possible model, influenced by the study of domestication, is that strong selection might fix through loci of major effect that themselves have strong pleiotropic effects (e.g. Clark et al. 2006; Stern and Orgogozo 2008) or that drag along allelic changes in loci that are physically linked to the target of selection (e.g. Olsen et al. 2006). It is unclear whether nature selects as monotonically strongly and/or is as forgiving of pleiotropic consequences (e.g. Westerbergh and Doebley 2002). But if natural were like artificial selection, it would suggest a role for large regulatory changes with compensatory mutations thereafter. The ability to generate detailed mechanistic information in crop systems, and extend this to the study of wild species, promises data that are essential for addressing these fundamental questions about the nature, pace, and comparability of selection by humans and in the wild.
Conclusions
Here we have discussed adaptive stress responses (specifically to water stress) as an example of potentially fruitful research reciprocity, where agricultural research has clearly produced advances that could benefit evolutionary studies, while evolutionary studies offer approaches and insights underexplored in crop studies. Although the emphasis here has been on one specific abiotic environmental response in one biological system, equivalent insights can be gained from integrating diverse approaches to understand traits mediating other abiotic and biotic interactions, and in other biological systems (Karrenberg and Widmer 2008; Alonso-Blanco et al. 2009).
Of course, the value of taking an intersecting approach to understanding agricultural and adaptive processes has been appreciated for decades. Sewell Wright, a pioneer of the modern synthesis, routinely published in journals that had both ‘pure’ and applied audiences, recognizing the plural application of evolutionary and genetic breeding principles. Throughout his storied career as a tomato breeder, Charles Rick publishing detailed papers on the pattern and distribution of natural genetic variation in wild tomato species, and argued vociferously for the value of natural variation for understanding and improving plant breeding (e.g. Rick 1978). In many ways, this advocacy has been responsible for the sustained interest in wild germplasm as a source of valuable genetic traits in tomato breeding, and the development of many resources to access this variation (Bai and Lindhout 2007).
Nonetheless, the increasing intersection of agricultural and evolutionary research interests has undoubtedly been facilitated by the recent and massive increase in the amount of genetic (genomic) information available in major crop and other model systems (Lister et al. 2009). Importantly, much of the value of these emerging genomic analyses has been rooted in the ability to undertake comparative ‘evolutionary’ analyses with closely related nonmodel relatives, whether these are as simple as pairwise comparisons of sequences or as comprehensive as population genomic studies of important trait variation (Storz 2005; Ehrenreich and Purugganan 2006; Begun et al. 2007; Dalziel et al. 2009). This continuing ‘evolutionary’ emphasis on incorporating diverse natural variation promises to offer unprecedented insight into molecular genetic mechanisms of agriculturally relevant trait variation (Ehrenreich and Purugganan 2006; Alonso-Blanco et al. 2009), and to allow the integration of functional, molecular, and evolutionary studies to understand the mechanistic basis of adaptive evolutionary change (Feder and Mitchell-Olds 2003; Vasemägi and Primmer 2005). By facilitating contemporary interchanges between applied and evolutionary research, these genomic advances have also cast a brighter light on the reciprocal lessons that can be learned from comparing and extending results in these two research traditions. Overall, combining the mechanistic insight of functional agricultural research with the theoretical and conceptual framework of evolutionary biology, promises to offer deeper insights into the genetic changes that are responsible for adaptive trait evolution. By capitalizing on these reciprocal insights, we can better address questions that are fundamental both to agriculture and evolution.
Acknowledgments
This work was supported in part by NSF grant DEB-0841957 to LCM; CDM is supported by an NSF Predoctoral Fellowship. We thank T. Nakazato and D. L. Warren for input into our ideas on ecological adaptation in wild tomatoes.
Literature cited
- Ackerly DD, Dudley SA, Sultan SE, Schmitt J, Coleman JS, Linder CR, Sandquist DR, et al. The evolution of plant ecophysiological traits: recent advances and future directions. BioScience. 2000;50:979–995. [Google Scholar]
- Alonso-Blanco C, Mendez-Vigo B, Koornneef M. From phenotypic to molecular polymorphisms involved in naturally occurring variation of plant development. International Journal of Developmental Biology. 2005;49:717–732. doi: 10.1387/ijdb.051994ca. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Aarts MGM, Bentsink L, Keurentjes JJB, Reymond M, Vreugdenhil D, Koornneef M. What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell. 2009;21:1877–1896. doi: 10.1105/tpc.109.068114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Ponce D, Aguade M, Rozas J. Network-level molecular evolutionary analysis of the insulin/TOR signal transduction pathway across 12 Drosophila genomes. Genome Research. 2009;19:234–242. doi: 10.1101/gr.084038.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araus JL, Slafer GA, Reynolds MP, Royo C. Plant breeding and drought in C-3 cereals: What should we breed for? Annals of Botany. 2002;89:925–940. doi: 10.1093/aob/mcf049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arntz AM, Delph LF. Pattern and process: evidence for the evolution of photosynthetic traits in natural populations. Oecologia. 2001;127:455–467. doi: 10.1007/s004420100650. [DOI] [PubMed] [Google Scholar]
- Atherton JG, Rudich J. The tomato crop: a scientific basis for improvement, pages 661. London: Chapman & Hall; 1986. [Google Scholar]
- Bai YL, Lindhout P. Domestication and breeding of tomatoes: What have we gained and what can we gain in the future? Annals of Botany. 2007;100:1085–1094. doi: 10.1093/aob/mcm150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone A, Di Matteo A, Carputo D, Frusciante L. High-throughput genomics enhances tomato breeding efficiency. Current Genomics. 2009;10:1–9. doi: 10.2174/138920209787581226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels D, Sunkar R. Drought and salt tolerance in plants. Critical Reviews in Plant Sciences. 2005;24:23–58. [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, Hillier LW, Poh YP, Hahn MW, Nista PM, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biology. 2007;5:2534–2559. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom AJ, Zwieniecki MA, Passioura JB, Randall LB, Holbrook NM, St Clair DA. Water relations under root chilling in a sensitive and tolerant tomato species. Plant, Cell and Environment. 2004;27:971–979. [Google Scholar]
- Bohnert HJ, Nelson DE, Jensen RG. Adaptations to environmental stresses. Plant Cell. 1995;7:1099–1111. doi: 10.1105/tpc.7.7.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer JS. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- Bray EA. Plant responses to water deficit. Trends in Plant Science. 1997;2:48–54. [Google Scholar]
- Brown JH. Macroecology: progress and prospect. Oikos. 1999;87:3–14. [Google Scholar]
- Caicedo AL, Schaal BA. Heterogeneous evolutionary processes affect R gene diversity in natural populations of Solanum pimpinellifolium. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17444–17449. doi: 10.1073/pnas.0407899101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G, Shapiro MD, Brady SD, Southwick AM, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin FS, Autumn K, Pugnaire F. Evolution of suites of traits in response to environmental stress. American Naturalist. 1993;142:S78–S92. [Google Scholar]
- Chaves MM, Oliveira MM. Mechanisms underlying plant resilience to water deficits: prospects for water-saving agriculture. Journal of Experimental Botany. 2004;55:2365–2384. doi: 10.1093/jxb/erh269. [DOI] [PubMed] [Google Scholar]
- Chen KY, Cong B, Wing R, Vrebalov J, Tanksley SD. Changes in regulation of a transcription factor lead to autogamy in cultivated tomatoes. Science. 2007;318:643–645. doi: 10.1126/science.1148428. [DOI] [PubMed] [Google Scholar]
- Chen GX, Krugman T, Fahima T, Chen KG, Hu YG, Roder M, Nevo E, et al. Chromosomal regions controlling seedling drought resistance in Israeli wild barley, Hordeum spontaneum C. Koch. Genetic Resources and Crop Evolution. 2010;57:85–99. [Google Scholar]
- Chetelat RT, Ji Y. Cytogenetics and evolution. In: Razdan MK, Mattoo AK, editors. Genetic Improvement of Solanaceous Crops. Enfield, NH: Science Publishers; 2007. pp. 77–112. [Google Scholar]
- Clark RM, Wagler TN, Quijada P, Doebley J. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nature Genetics. 2006;38:594–597. doi: 10.1038/ng1784. [DOI] [PubMed] [Google Scholar]
- Collins NC, Tardieu F, Tuberosa R. Quantitative trait loci and crop performance under abiotic stress: Where do we stand? Plant Physiology. 2008;147:469–486. doi: 10.1104/pp.108.118117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock JP, McCouch SR, Martin BC, Tauer CG, Vision TJ, Xu YB, Pausch RC. The effects of resource availability and environmental conditions on genetic rankings for carbon isotope discrimination during growth in tomato and rice. Functional Plant Biology. 2005;32:1089–1105. doi: 10.1071/FP05117. [DOI] [PubMed] [Google Scholar]
- Condon AG, Richards RA, Rebetzke GJ, Farquhar GD. Breeding for high water-use efficiency. Journal of Experimental Botany. 2004;55:2447–2460. doi: 10.1093/jxb/erh277. [DOI] [PubMed] [Google Scholar]
- Cutler AJ. Abscisic Acid (ABA) Encyclopedia of Life Sciences (ELS) Chichester: John Wiey & Sons; 2009. [Google Scholar]
- Dalziel AC, Rogers SM, Schulte PM. Linking genotypes to phenotypes and fitness: how mechanistic biology can inform molecular ecology. Molecular Ecology. 2009;18:4997–5017. doi: 10.1111/j.1365-294X.2009.04427.x. [DOI] [PubMed] [Google Scholar]
- D'Arcy WG. The classification of the Solanaceae. In: Hawkes JG, Lester RN, Skelding AD, editors. The Biology and Taxonomy of the Solanaceae. London: Academic Press; 1979. pp. 3–47. [Google Scholar]
- Darwin C. On the Origin of Species by Means of Natural Selection. London: John Murray; 1859. [Google Scholar]
- Dudley SA. Differing selection on plant physiological traits in response to environmental water availability: a test of adaptive hypotheses. Evolution. 1996a;50:92–102. doi: 10.1111/j.1558-5646.1996.tb04475.x. [DOI] [PubMed] [Google Scholar]
- Dudley SA. The response to differing selection on plant physiological traits: evidence for local adaptation. Evolution. 1996b;50:103–110. doi: 10.1111/j.1558-5646.1996.tb04476.x. [DOI] [PubMed] [Google Scholar]
- Eanes WF. Analysis of selection on enzyme polymorphisms. Annual Review of Ecology and Systematics. 1999;30:301–326. [Google Scholar]
- Easlon HM, Richards JH. Drought response in self-compatible species of tomato (Solanaceae) American Journal of Botany. 2009;96:605–611. doi: 10.3732/ajb.0800189. [DOI] [PubMed] [Google Scholar]
- Ehleringer JR, Monson RK. Evolutionary and ecological aspects of photosynthetic pathway variation. Annual Review of Ecology and Systematics. 1993;24:411–439. [Google Scholar]
- Ehrenreich IM, Purugganan MD. The molecular genetic basis of plant adaptation. American Journal of Botany. 2006;93:953–962. doi: 10.3732/ajb.93.7.953. [DOI] [PubMed] [Google Scholar]
- Feder ME, Mitchell-Olds T. Evolutionary and ecological functional genomics. Nature Reviews Genetics. 2003;4:651–657. doi: 10.1038/nrg1128. [DOI] [PubMed] [Google Scholar]
- Fernandez AI, Viron N, Alhagdow M, Karimi M, Jones M, Amsellem Z, Sicard A, et al. Flexible tools for gene expression and silencing in Tomato. Plant Physiology. 2009;151:1729–1740. doi: 10.1104/pp.109.147546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Tadmor Y, Zamir D. Natural genetic variation for improving crop quality. Current Opinion in Plant Biology. 2006;9:196–202. doi: 10.1016/j.pbi.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The Genetical Theory of Natural Selection. Oxford: Clarendon Press; 1930. [Google Scholar]
- Flowers JM, Sezgin E, Kumagai S, Duvernell DD, Matzkin LM, Schmidt PS, Eanes WF. Adaptive evolution of metabolic pathways in Drosophila. Molecular Biology and Evolution. 2007;24:1347–1354. doi: 10.1093/molbev/msm057. [DOI] [PubMed] [Google Scholar]
- Flowers JM, Hanzawa Y, Hall MC, Moore RC, Purugganan MD. Population genomics of the Arabidopsis thaliana flowering time gene network. Molecular Biology and Evolution. 2009;26:2475–2486. doi: 10.1093/molbev/msp161. [DOI] [PubMed] [Google Scholar]
- Foolad MR. Genome mapping and molecular breeding of tomato. International Journal of Plant Genomics. 2007;2007:64358. doi: 10.1155/2007/64358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foolad MR, Zhang LP, Subbiah P. Genetics of drought tolerance during seed germination in tomato: inheritance and QTL mapping. Genome. 2003;46:536–545. doi: 10.1139/g03-035. [DOI] [PubMed] [Google Scholar]
- Frankel N, Hasson E, Iusem ND, Rossi MS. Adaptive evolution of the water stress-induced gene Asr2 in Lycopersicon species dwelling in arid habitats. Molecular Biology and Evolution. 2003;20:1955–1962. doi: 10.1093/molbev/msg214. [DOI] [PubMed] [Google Scholar]
- Frary A, Xu YM, Liu JP, Mitchell S, Tedeschi E, Tanksley S. Development of a set of PCR-based anchor markers encompassing the tomato genome and evaluation of their usefulness for genetics and breeding experiments. Theoretical and Applied Genetics. 2005;111:291–312. doi: 10.1007/s00122-005-2023-7. [DOI] [PubMed] [Google Scholar]
- Gay AP, Hurd RG. Influence of light on stomatal density in tomato. New Phytologist. 1975;75:37–46. [Google Scholar]
- Geber MA, Griffen LR. Inheritance and natural selection on functional traits. International Journal of Plant Sciences. 2003;164:S21–S42. [Google Scholar]
- Giombini MI, Frankel N, Iusem ND, Hasson E. Nucleotide polymorphism in the drought responsive gene Asr2 in wild populations of tomato. Genetica. 2008;136:13–25. doi: 10.1007/s10709-008-9295-1. [DOI] [PubMed] [Google Scholar]
- Gur A, Zamir D. Unused natural variation can lift yield barriers in plant breeding. PLoS Biology. 2004;2:1610–1615. doi: 10.1371/journal.pbio.0020245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. Distinguishing among evolutionary models for the maintenance of gene duplicates. Journal of Heredity. 2009;100:605–617. doi: 10.1093/jhered/esp047. [DOI] [PubMed] [Google Scholar]
- Hancock JF. Contributions of domesticated plant studies to our understanding of plant evolution. Annals of Botany. 2005;96:953–963. doi: 10.1093/aob/mci259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey PH, Pagel MD. The Comparative Method in Evolutionary Biology: Oxford Series in Evolutionary Biology. Oxford: Oxford University Press; 1991. [Google Scholar]
- Hereford J. A quantitative survey of local adaptation and fitness trade-offs. American Naturalist. 2009;173:579–588. doi: 10.1086/597611. [DOI] [PubMed] [Google Scholar]
- Heschel MS, Donohue K, Hausmann N, Schmitt J. Population differentiation and natural selection for water-use efficiency in Impatiens capensis (Balsaminaceae) International Journal of Plant Sciences. 2002;163:907–912. [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: Evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Hsiao TC. Plant responses to water stress. Annual Review of Plant Physiology and Plant Molecular Biology. 1973;24:519–570. [Google Scholar]
- Jimenez-Gomez JM, Maloof JN. Sequence diversity in three tomato species: SNPs, markers, and molecular evolution. BMC Plant Biology. 2009;9:85. doi: 10.1186/1471-2229-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johal GS, Balint-Kurti P, Well CF. Mining and harnessing natural variation: a little MAGIC. Crop Science. 2008;48:2066–2073. [Google Scholar]
- Kacser H, Burns JA. The control of flux. Symposia of the Society for Experimental Biology. 1973;27:65–104. [PubMed] [Google Scholar]
- Kahn TL, Fender SE, Bray EA, Oconnell MA. Characterization of expression of drought and abscisic acid-regulated tomato genes in the drought-resistant species Lycopersicon pennellii. Plant Physiology. 1993;103:597–605. doi: 10.1104/pp.103.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenetzky L, Asis R, Bassi S, De Godoy F, Bermudez L, Fernie AR, Van Sluys MA, et al. Genomic analysis of wild tomato (Solanum pennellii) introgression determining metabolic- and yield-associated traits. Plant Physiology. 2010;152:1772–1786. doi: 10.1104/pp.109.150532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrenberg S, Widmer A. Ecologically relevant genetic variation from a non-Arabidopsis perspective. Current Opinion In Plant Biology. 2008;11:156–162. doi: 10.1016/j.pbi.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Kebede H, Martin B, Nienhuis J, King G. Leaf anatomy of 2 Lycopersicon species with contrasting gas-exchange properties. Crop Science. 1994;34:108–113. [Google Scholar]
- Kopp A. Metamodels and phylogenetic replication: a systematic approach to the evolution of developmental pathways. Evolution. 2009;63:2771–2789. doi: 10.1111/j.1558-5646.2009.00761.x. [DOI] [PubMed] [Google Scholar]
- Kozak KH, Graham CH, Wiens JJ. Integrating GIS-based environmental data into evolutionary biology. Trends in Ecology & Evolution. 2008;23:141–148. doi: 10.1016/j.tree.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Lambers H, Chapin FS, Pons TL. Plant Physiological Ecology. New York: Springer; 2008. [Google Scholar]
- Latta RG. Testing for local adaptation in Avena barbata: a classic example of ecotypic divergence. Molecular Ecology. 2009;18:3781–3791. doi: 10.1111/j.1365-294X.2009.04302.x. [DOI] [PubMed] [Google Scholar]
- Lippman ZB, Semel Y, Zamir D. An integrated view of quantitative trait variation using tomato interspecific introgression lines. Current Opinion in Genetics & Development. 2007;17:545–552. doi: 10.1016/j.gde.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Lister R, Gregory BD, Ecker JR. Next is now: new technologies for sequencing of genomes, transcriptomes, and beyond. Current Opinion in Plant Biology. 2009;12:107–118. doi: 10.1016/j.pbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow MM. Strategies of response to water stress. In: Kreeb KH, Richter H, Hinckley TM, editors. Structural and Functional Responses to Environmental Stresses. The Hague: SPB Academic; 1989. pp. 269–281. [Google Scholar]
- Maldonado C, Squeo FA, Ibacache E. Phenotypic response of Lycopersicon chilense to water deficit. Revista Chilena De Historia Natural. 2003;76:129–137. [Google Scholar]
- Martin B, Thorstenson YR. Stable carbon isotope composition (delta-C-13), water-use efficiency, and biomass productivity of Lycopersicon esculentum Lycopersicon pennellii, and the F1 hybrid. Plant Physiology. 1988;88:213–217. doi: 10.1104/pp.88.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B, Nienhuis J, King G, Schaefer A. Restriction fragment length polymorphisms associated with water-use efficiency in tomato. Science. 1989;243:1725–1728. doi: 10.1126/science.243.4899.1725. [DOI] [PubMed] [Google Scholar]
- Martin B, Tauer CG, Lin RK. Carbon isotope discrimination as a tool to improve water-use efficiency in tomato. Crop Science. 1999;39:1775–1783. [Google Scholar]
- McKay JK, Stinchcombe JR. Ecological genomics of model eukaryotes. Evolution. 2008;62:2953–2957. doi: 10.1111/j.1558-5646.2008.00536.x. [DOI] [PubMed] [Google Scholar]
- McKay JK, Richards JH, Mitchell-Olds T. Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotropy contributes to genetic correlations among ecological traits. Molecular Ecology. 2003;12:1137–1151. doi: 10.1046/j.1365-294x.2003.01833.x. [DOI] [PubMed] [Google Scholar]
- McKay JK, Richards JH, Nemali KS, Sen S, Mitchell-Olds T, Boles S, Stahl EA, et al. Genetics of drought adaptation in Arabidopsis thaliana II. QTL analysis of a new mapping population, KAS-1 x TSU–1. Evolution. 2008;62:3014–3026. doi: 10.1111/j.1558-5646.2008.00474.x. [DOI] [PubMed] [Google Scholar]
- Mitchell JP, Shennan C, Grattan SR, May DM. Tomato fruit yields and quality under water deficit and salinity. Journal of the American Society for Horticultural Science. 1991;116:215–221. [Google Scholar]
- Moyle LC. Ecological and evolutionary genomics in the wild tomatoes (Solanum Sect. Lycopersicon) Evolution. 2008;62:2995–3013. doi: 10.1111/j.1558-5646.2008.00487.x. [DOI] [PubMed] [Google Scholar]
- Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, Lin CW, et al. The SOL Genomics Network. A comparative resource for Solanaceae biology and beyond. Plant Physiology. 2005;138:1310–1317. doi: 10.1104/pp.105.060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir CD, Moyle LC. Antagonistic epistasis for ecophysiological trait differences between Solanum species. New Phytologist. 2009;183:789–802. doi: 10.1111/j.1469-8137.2009.02949.x. [DOI] [PubMed] [Google Scholar]
- Nakazato T, Bogonovich M, Moyle LC. Environmental factors predict adaptive phenotypic differentiation within and between two wild andean tomatoes. Evolution. 2008;62:774–792. doi: 10.1111/j.1558-5646.2008.00332.x. [DOI] [PubMed] [Google Scholar]
- Nakazato T, Warren DL, Moyle LC. Ecological and geographic modes of species divergence in wild tomatoes. American Journal of Botany. 2010;97:680–693. doi: 10.3732/ajb.0900216. [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- Nesbitt TC, Tanksley SD. Comparative sequencing in the genus Lycopersicon: implications for the evolution of fruit size in the domestication of cultivated tomatoes. Genetics. 2002;162:365–379. doi: 10.1093/genetics/162.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo E, Chen GX. Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant Cell and Environment. 2010;33:670–685. doi: 10.1111/j.1365-3040.2009.02107.x. [DOI] [PubMed] [Google Scholar]
- Nielsen R. Molecular signatures of natural selection. Annual Review of Genetics. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- Oakley TH. Today's multiple choice exam: (a) gene duplication; (b) structural mutation; (c) co-option; (d) regulatory mutation; (e) all of the above. Evolution & Development. 2007;9:523–524. doi: 10.1111/j.1525-142X.2007.00192.x. [DOI] [PubMed] [Google Scholar]
- Olsen KM, Caicedo AL, Polato N, McClung A, McCouch S, Purugganan MD. Selection under domestication: evidence for a sweep in the rice Waxy genomic region. Genetics. 2006;173:975–983. doi: 10.1534/genetics.106.056473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. The genetic theory of adaptation: a brief history. Nature Reviews Genetics. 2005;6:119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- Passioura JB. Drought and drought tolerance. Plant Growth Regulation. 1996;20:79–83. [Google Scholar]
- Patterson BD, Paull R, Smillie RM. Chilling resistance in Lycopersicon hirsutum Humb. and Bonpl. a wild tomato with a wide altitudinal distribution. Australian Journal of Plant Physiology. 1978;5:609–617. [Google Scholar]
- Peralta IE, Spooner DM. Granule-bound starch synthase (GBSSI) gene phylogeny of wild tomatoes (Solanum L. section Lycopersicon [Mill.] Wettst. subsection Lycopersicon) American Journal of Botany. 2001;88:1888–1902. [PubMed] [Google Scholar]
- Peralta IE, Spooner DM, Knapp S. Taxonomy of wild tomatoes and their relatives (Solanum sect Lycopersicoides, sect. Juglandifolia, sect. Lycopersicon; Solanaceae) Systematic Botany Monographs. 2008;84:1–186. [Google Scholar]
- Phillips PC. Epistasis – the essential role of gene interactions in the structure and evolution of genetic systems. Nature Reviews Genetics. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan MD, Fuller DQ. The nature of selection during plant domestication. Nature. 2009;457:843–848. doi: 10.1038/nature07895. [DOI] [PubMed] [Google Scholar]
- Quiros CF. Lycopersicon cytogenetics. In: Gupta PK, Tsuchiya T, editors. Chromosome Engineering in Plants: Genetics, Breeding, Evolution. Amsterdam: Elsevier; 1991. pp. 119–137. [Google Scholar]
- Ramsay H, Rieseberg LH, Ritland K. The correlation of evolutionary rate with pathway position in plant terpenoid biosynthesis. Molecular Biology and Evolution. 2009;26:1045–1053. doi: 10.1093/molbev/msp021. [DOI] [PubMed] [Google Scholar]
- Rausher MD. Evolutionary transitions in floral color. International Journal of Plant Sciences. 2008;169:7–21. [Google Scholar]
- Reid JB, Renquist AR. Enhanced root production as a feed-forward response to soil water deficit in field-grown tomatoes. Australian Journal of Plant Physiology. 1997;24:685–692. [Google Scholar]
- Renquist AR, Reid JB. Processing tomato fruit quality: influence of soil water deficits at flowering and ripening. Australian Journal of Agricultural Research. 2001;52:793–799. [Google Scholar]
- Richards RA. Defining selection criteria to improve yield under drought. Plant Growth Regulation. 1996;20:157–166. [Google Scholar]
- Rick CM. Potential genetic resources in tomato species: clues from observations in native habitats. In: Srb AM, editor. Genes, Enzymes, and Populations. New York, NY: Plenum Press; 1973. pp. 255–269. [DOI] [PubMed] [Google Scholar]
- Rick CM. Natural variability in wild species of Lycopersicon and its bearing on tomato breeding. Agraria. 1976;30:249–259. [Google Scholar]
- Rick CM. Tomato Germplasm Resources. Shanhua, Taiwan: The 1st International Symposium on Tropical Tomato; 1978. pp. 214–224. [Google Scholar]
- Rick CM. Biosystematic studies in Lycopersicon and closely related species of Solanum. In: Hawkes JG, Lester RN, Skelding AD, editors. The Biology and Taxonomy of the Solanaceae. Vol. 7. London: Academic Press; 1979. pp. 667–679. [Google Scholar]
- Romero-Aranda R, Soria T, Cuartero J. Tomato plant-water uptake and plant-water relationships under saline growth conditions. Plant Science. 2001;160:265–272. doi: 10.1016/s0168-9452(00)00388-5. [DOI] [PubMed] [Google Scholar]
- Rose LE, Michelmore RW, Langley CH. Natural variation in the Pto disease resistance gene within species of wild tomato (Lycopersicon). II. Population genetics of Pto. Genetics. 2007;175:1307–1319. doi: 10.1534/genetics.106.063602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Ibarra J, Morrell PL, Gaut BS. Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8641–8648. doi: 10.1073/pnas.0700643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudich J, Kalmar D, Geizenberg C, Harel S. Low water tensions in defined growth stages of processing tomato plants and their effects on yield and quality. Journal of Horticultural Science. 1977;52:391–399. [Google Scholar]
- Stebbins GL. Aridity as a stimulus to plant evolution. American Naturalist. 1952;86:33–44. [Google Scholar]
- Stern DL, Orgogozo V. The loci of evolution: How predictable is genetic evolution? Evolution. 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens MA, Rudich J. Genetic potential for overcoming physiological limitations on adaptability, yield, and quality in the tomato. Hortscience. 1978;13:673–678. [Google Scholar]
- Storz JF. Using genome scans of DNA polymorphism to infer adaptive population divergence. Molecular Ecology. 2005;14:671–688. doi: 10.1111/j.1365-294X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Sultan SE. Development in context: the timely emergence of eco-devo. Trends in Ecology & Evolution. 2007;22:575–582. doi: 10.1016/j.tree.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Tan CS. Tomato yield-evapotranspiration relationships, seasonal canopy temperature and stomatal conductance as affected by irrigation. Canadian Journal of Plant Science. 1993;73:257–264. [Google Scholar]
- Thakur PS. Different physiological-responses of tomato (Lycopersicon-Esculentum Mill) cultivars to drought. Acta Physiologiae Plantarum. 1990;12:175–182. [Google Scholar]
- Thompson AJ, Jackson AC, Parker RA, Morpeth DR, Burbidge A, Taylor IB. Abscisic acid biosynthesis in tomato: regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Molecular Biology. 2000;42:833–845. doi: 10.1023/a:1006448428401. [DOI] [PubMed] [Google Scholar]
- Thompson AJ, Thorne ET, Burbidge A, Jackson AC, Sharp RE, Taylor IB. Complementation of notabilis, an abscisic acid-deficient mutant of tomato: importance of sequence context and utility of partial complementation. Plant, Cell and Environment. 2004;27:459–471. [Google Scholar]
- Thompson AJ, Mulholland BJ, Jackson AC, McKee JMT, Hilton HW, Symonds RC, Sonneveld T, et al. Regulation and manipulation of ABA biosynthesis in roots. Plant, Cell and Environment. 2007a;30:67–78. doi: 10.1111/j.1365-3040.2006.01606.x. [DOI] [PubMed] [Google Scholar]
- Thompson AJ, Andrews J, Mulholland BJ, McKee JMT, Hilton HW, Black CR, Taylor IB. Overproduction of abscisic acid in tomato increases transpiration efficiency and root hydraulic conductivity and influences leaf expansion. Plant Physiology. 2007b;143:1905–1917. doi: 10.1104/pp.106.093559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticha I. Photosynthetic characteristics during ontogenesis of leaves.7. Stomata density and sizes. Photosynthetica. 1982;16:375–471. [Google Scholar]
- Torrecillas A, Guillaume C, Alarcon JJ, Ruizsanchez MC. Water relations of 2 tomato species under water-stress and recovery. Plant Science. 1995;105:169–176. [Google Scholar]
- Vallejos CE. Genetic diversity of plants for response to low temperatures and its potential use in crop plants. In: Lyons JM, Graham D, Raison JK, editors. Low Temperature Stress in Crop Plants: The Role of the Membrane. New York, NY: Academic Press; 1979. p. 565. [Google Scholar]
- Van Deynze A, Stoffel K, Buell CR, Kozik A, Liu J, Van Der Knaap E, Francis D. Diversity in conserved genes in tomato. BMC Genomics. 2007;8:465. doi: 10.1186/1471-2164-8-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasemägi A, Primmer CR. Challenges for identifying functionally important genetic variation: the promise of combining complementary research strategies. Molecular Ecology. 2005;14:3623–3642. doi: 10.1111/j.1365-294X.2005.02690.x. [DOI] [PubMed] [Google Scholar]
- Weinig C, Schmitt J. Environmental effects on the expression of quantitative trait loci and implications for phenotypic evolution. BioScience. 2004;54:627–635. [Google Scholar]
- Westerbergh A, Doebley J. Morphological traits defining species differences in wild relatives of maize are controlled by multiple quantitative trait loci. Evolution. 2002;56:273–283. doi: 10.1111/j.0014-3820.2002.tb01337.x. [DOI] [PubMed] [Google Scholar]
- Wudiri BB, Henderson DW. Effects of water-stress on flowering and fruit-set in processing tomatoes. Scientia Horticulturae. 1985;27:189–198. [Google Scholar]
- Xu X, Martin B, Comstock JP, Vision TJ, Tauer CG, Zhao B, Pausch RC, et al. Fine mapping a QTL for carbon isotope composition in tomato. Theoretical and Applied Genetics. 2008;117:221–233. doi: 10.1007/s00122-008-0767-6. [DOI] [PubMed] [Google Scholar]
- Yoo CY, Pence HE, Hasegawa PM, Mickelbart MV. Regulation of transpiration to improve crop water use. Critical Reviews in Plant Sciences. 2009;28:410–431. [Google Scholar]
- Young KR, Ulloa CU, Luteyn JL, Knapp S. Plant evolution and endemism in Andean South America: an introduction. Botanical Review. 2002;68:4–21. [Google Scholar]
- Zamir D. Improving plant breeding with exotic genetic libraries. Nature Reviews Genetics. 2001;2:983–989. doi: 10.1038/35103590. [DOI] [PubMed] [Google Scholar]


