Abstract
Pathogenicity and genetic diversity of Fusarium oxysporum from geographically widespread native Gossypium populations, including a cotton growing area believed to be the center of origin of VCG 01111 and VCG 01112 of F. oxysporum f. sp. vasinfectum (Fov) in Australia, was determined using glasshouse bioassays and AFLPs. Five lineages (A–E) were identified among 856 isolates. Of these, 12% were strongly pathogenic on cotton, 10% were weakly pathogenic and designated wild Fov, while 78% were nonpathogenic. In contrast to the occurrence of pathogenic isolates in all five lineages in soils associated with wild Gossypium, in cotton growing areas only three lineages (A, B, E) occurred and all pathogenic isolates belonged to two subgroups in lineage A. One of these contained VCG 01111 isolates while the other contained VCG 01112 isolates. Sequence analyses of translation elongation factor-1α, mitochondrial small subunit rDNA, nitrate reductase and phosphate permease confirmed that Australian Fov isolates were more closely related to lineage A isolates of native F. oxysporum than to Fov races 1–8 found overseas. These results strongly support a local evolutionary origin for Fov in Australian cotton growing regions.
Keywords: agriculture, cotton, fungal, fusarium wilt, host shift, host–pathogen, pathogen emergence, plant disease
Introduction
Widespread and increasing human impacts on all levels of biological organization (e.g. changes in land-use patterns, fragmentation of natural ecosystems) suggests that there is value in the application of evolutionary principles to emerging issues relating to these processes (Thompson 2005). One clear example where this is of direct relevance is with regard to the incidence and prevalence of disease in agro-ecosystems, particularly in the context of the role that interactions between production and native components of these landscapes might play in the emergence and spread of new diseases (Anderson et al. 2004; Burdon and Thrall 2008). This is not only with respect to understanding the underlying epidemiological and evolutionary processes, but also with regard to identifying appropriate control strategies (e.g. Ewald 1994; Jeger et al. 2006; Gilligan 2008). Here, we present results from extensive studies of Fusarium wilt disease in Australian cotton growing regions, with the aim of evaluating likely evolutionary origins and agronomic management implications for the pathogen. Of particular note is the fact that there are a number of native Gossypium species in Australia, raising the possibility that these wild relatives have played a role in the evolution of the pathogen, as has been demonstrated in other systems (e.g. Burdon et al. 1983; Oates et al. 1983; Frenkel et al. 2007).
Fusarium oxysporum f. sp. vasinfectum (Fov) is a soil-borne fungal pathogen of cotton (Gossypium hirsutum L.) characterized by a parasitic phase within the vascular tissue and a saprophytic phase in the soil or plant residue after host death. Worldwide, eight races have been characterized based on pathogenicity on differential host sets (Chen et al. 1985; Hillocks 1992), and 12 vegetative compatibility groups (VCGs) identified, each presumably representing a clonal lineage (Fernandez et al. 1994; Bentley et al. 2000). Genetic evidence has demonstrated that the eight pathogenic races of F. oxysporum f. sp. vasinfectum are polyphyletic with at least two independent evolutionary origins (Skovgaard et al. 2001).
In Australia, Fusarium wilt was first recorded in the Brookstead, Cecil Plains and Boggabilla regions of Queensland/New South Wales in 1993/1994 from which it subsequently spread to most major cotton growing regions. While Australian isolates of Fov are pathogenically similar to race 6 on the standard differential hosts, they belong to VC groups 01111 and 01112 which are vegetatively incompatible with all non-Australian isolates of the pathogen, including race 6 isolates (Davis et al. 1996; Bentley et al. 2000). Furthermore, phylogenetic analysis of multigene sequences and pathogenicity has shown that Australian Fov are distinct from all races and VCGs found in California and China (Kim et al. 2005).
In Australia, the two VCGs have different geographic distributions with VCG 01111 occurring in all infected cotton growing regions, while VCG 01112 is restricted to the Boggabilla region where it was first detected (Wang et al. 2006). Despite genetic variation among isolates, no clear spatial population structure has been found at the largest spatial scale. However, both the greatest genetic diversity and some indication of local population differentiation was observed in the Boggabilla region, which, when coupled with the first reports of Fusarium wilt of cotton originating from this area (Kochman 1995), suggests that this may be the centre of origin of the two VCGs of Fov in Australia (Wang et al. 2006).
Understanding the evolutionary origin of new pathogens is important for effective disease management as strategies to control introduced pathogens may differ from those for pathogens that arise locally. New occurrences of Fusarium wilt pathogens are frequently the result of recent introductions rather than independent local origins (Gordon and Martyn 1997), but the simultaneous appearance of two distinct VCGs of F. oxysporum f. sp. vasinfectum suggests that this may not be the case in Australia. Given the clonal nature of Fov, distinguishing between long-distance migration and local evolution as a source of origin should be relatively straightforward. An introduced pathogen is likely to be genetically distinct from the pre-existing pool of local F. oxysporum isolates, while a locally derived pathogen should be more closely related to sympatric nonpathogen types.
Cotton was introduced to Australia with European settlement in 1788, but not grown extensively until the early 1960s. However, 17 wild Gossypium species are indigenous to Australia, four of which (G. australe, G. bickii, G. nelsonii, G. sturtianum) have native ranges that overlap or abut areas where the majority of cultivated cotton is grown (Craven et al. 1994). Interestingly, a survey of Fusarium species associated with these wild cottons detected a number of F. oxysporum isolates that caused mild, but typical, foliar and vascular symptoms of Fusarium wilt on cultivated cotton (Wang et al. 2004), which suggests that Fov may have existed in Australia before cotton was introduced. This raises the possibility that the two Fov VCGs found in commercial cotton fields evolved locally. Such evolution of pathogenicity in F. oxysporum has previously been documented in other crops including melon and tomato (Katan et al. 1994; Rosewich et al. 1999; Cai et al. 2003).
Uncultivated areas within agricultural production systems may represent reservoirs of native microflora similar to those that would have been present in adjacent agricultural soils prior to cultivation (Gordon et al. 1992). This suggests that if a new crop pathogen arises in situ, it is likely to show close relatedness to nonpathogenic isolates occurring in such nearby uncultivated areas. For example, a local origin for Fusarium root rot of pea in Denmark was implicated by the close DNA sequence homology of pathogenic strains with nonpathogenic isolates collected from the same fields (Skovgaard et al. 2002).
The primary goal of this study was to assess the hypothesis that VCG 01111 and VCG 01112 of F. oxysporum f. sp. vasinfectum evolved from local F. oxysporum populations in Australia. To do this, we determined genetic relationships between Fov isolates found in cotton fields, in nearby uncultivated soils, and indigenous F. oxysporum isolates found in a range of soils associated with native Gossypium species.
Materials and methods
Reference isolates of Australian F. oxysporum f. sp. vasinfectum
Isolates 24500 and 24595 of VCG 01111 and isolates 24492 and B/96/02 of VCG 01112, provided by Natalie Moore and Wayne O'Neil (Queensland Department of Primary Industries, Indooroopilly, Australia), were used as references of pathogenic Australian Fov in this study.
Sample collection
Soil was collected from a total of 90 populations of four native Gossypium species (G. australe, G. bickii, G. nelsonii, G. sturtianum) in 2001–2002 in the eastern and central parts of Australia (Table 1). At each site c. 200 g of soil was taken from the rhizosphere of 3–10 plants after the surface 2 cm layer was removed.
Table 1.
Number of Gossypium populations sampled in this study and incidence of Fusarium oxysporum and wild F. oxysporum f. sp. vasinfectum (Fov) in populations summarized by Gossypium species and geographic regions respectively
| Sources of populations | Number of populations sampled | Number (%)* of populations associated with F. oxysporum | Number (%) of populations associated with wild Fov |
|---|---|---|---|
| By Gossypium species | |||
| G. australe | 33 | 16 (48) | 11 (33) |
| G. bickii | 13 | 8 (62) | 4 (31) |
| G. nelsonii | 11 | 6 (55) | 4 (36) |
| G. sturtianum | 33 | 30 (91) | 20 (61) |
| By geographic regions | |||
| Mount Isa (QLD) 20°15′–32°05′S; 139°00′–150°59′E | 14 | 8 (57) | 4 (29) |
| Longreach-Theodore (QLD) 20°15′–32°05′S; 139°00′–150°59′E | 12 | 11 (92) | 8 (67) |
| Alice Springs-Tennant Creek (NT) 19°17′–23°49′S; 132°44′–138°00′E | 51 | 30 (59) | 17 (33) |
| Leigh Creek-Arkaroola (SA) 30°00′–31°02′S; 137°46′–139°26′E | 13 | 11 (85) | 10 (77) |
| Total | 90 | 60 (67) | 39 (43) |
Percentage of the populations in the total sampled.
Within the cotton growing region, 200 g of soil was collected from each of five randomly chosen points in an uncultivated plot of native vegetation in the Boggabilla region in 2002 (Fig. 1). This site comprised a fenced minimally disturbed grassy woodland of c. 1.5 km2 that had never been cultivated.
Figure 1.
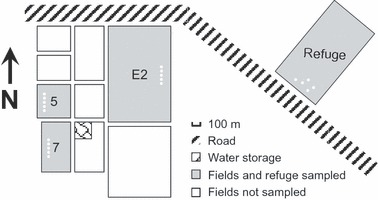
Diagram showing spatial relationships among the uncultivated land (refuge) and the three cultivated fields sampled in the Boggabilla region of New South Wales. White spots in the fields and refuge indicate the source of the plant and soil samples.
Soil and plant samples were collected in 2002 and 2004 from three cultivated cotton crops in fields (7, 5, and E2; Fig. 1) in which cotton had been grown in wheat or fallow rotation since the 1980s. These fields were all within c. 1 km of the native vegetation site. In each field, 200 g of soil was collected from each of five positions that were >50 m from field margins and 10 rows apart. At the same time, 20–35 symptomatic plants were randomly sampled in the same fields by cutting a 10-cm stem section from the main shoot. Both soil and plant samples were air-dried at ambient temperature. Soils were ground, passed through a 710-μm sieve and then stored at 4°C until assayed.
Fungal isolation
Isolation was conducted on Peptone PCNB agar (Burgess et al. 1994). For isolation of F. oxysporum from soils, 0.5 g of soil was sprinkled onto five plates and incubated at 25°C for 1 week. All colonies morphologically resembling F. oxysporum were re-grown from single spores and subcultured. Finally, isolates were grown on carnation leaf agar plates at 25°C with a 12-h photoperiod, and F. oxysporum identified following Leslie and Summerell (2006). For isolation of Fov from plant samples, stem sections were surface sterilized in 0.5% sodium hypochlorite for 5 min and peeled under aseptic conditions. Small pieces of discoloured vascular tissue were placed on plates and incubated at 25°C for 1 week. Fungal hyphae growing out of tissue pieces were subcultured. The above procedure was repeated if F. oxysporum was not recovered in the initial attempt. Samples were considered free of F. oxysporum if both attempts were unsuccessful.
All isolates were grown on 10% potato dextrose agar (PDA; Difco Laboratories, Detroit, MI, USA) slants at 25°C for 1 week. Conidia were washed off by adding 1.5 mL of sterile 15% glycerol into each tube and pipetting the liquid several times. Conidial suspensions were stored in 2.0 mL cryogenic vials at −80°C.
Pathogenicity screening tests
Strains were tested for pathogenicity against a highly susceptible cotton cultivar, Siokra 1–4. Inoculum ranging in concentration from 2.5 × 105 to 8.5 × 107 spores/mL was prepared by growing strains on an orbital shaker in 75 mL of 25% potato dextrose broth (PDB; Difco) at 18–23°C for 1 week. Two-week-old seedlings were inoculated by dipping the roots in inocula for 5 min. Distilled water and a conidial suspension of Fov isolate 24500 (VCG 01111), were used as noninoculated and positive controls, respectively. Treated plants were transplanted into fresh potting mix (compost and perlite; 50/50, v/v) and grown at 18–23°C in a naturally lit glasshouse. A total of nine plants in three pots were challenged with each strain. Fusarium wilt was identified by the appearance of dark-brown discoloration in the vascular tissue and foliar necrosis 6 weeks after inoculation. Disease severity was assessed on a 0–4 scale (0 = asymptomatic; 1 = vascular discolouration only; 2 = necrosis on ≤50% of the foliage; 3 ≥ 50% but <100% foliar necrosis; 4 = 100% foliar necrosis).
The entire test was repeated and strains causing a mean disease severity of >1.5 were putatively identified as F. oxysporum f. sp. vasinfectum as suggested by Armstrong and Armstrong (1981). Isolates showing pathogenicity in both tests and producing a mean disease severity in the range of 0.1–1.5 were designated as wild Fov, i.e. weakly pathogenic on cotton.
Virulence comparison tests
The virulence (i.e. severity of disease symptoms) of wild Fov from soils associated with wild Gossypium populations and that found in cotton fields was compared on a moderately tolerant cotton cultivar Sicot 189 and a susceptible wild cotton (G. sturtianum, Gos-5250). Virulence comparison and pathogenicity screening tests used the same methodology except that in the former tests G. sturtianum seedlings were inoculated when 4 weeks old with a conidial suspension (1.0 ± 0.2 × 106 conidia/mL) from which hyphae had been removed by straining through tissue. All tests were conducted twice with three replicates for each strain. For each replicate, 30 plants were used in each trial involving cotton, but due to a lack of seeds only seven and nine plants, respectively, were used in the first and second trials involving G. sturtianum.
DNA extraction
Strains were grown for 3 days in 12 mL of 80% PDB in 15 mL sterile test tubes at 25°C after which mycelium was harvested by centrifuging cultures (2800 g for 15 min), decanting liquid, and transferring the pellet onto Whatman No.1 filter paper to remove excess water. Genomic DNA was extracted from lyophilized mycelia using DNeasy Plant kits (Qiagen Pty Ltd, Clifton Hill, Australia). DNA concentrations were determined using a GeneQuant II spectrophotometer (Pharmacia Biotech, Cambridge, England) and adjusted to 50 ng/μL.
AFLP analysis
AFLP fingerprints were generated using the protocol described by Vos et al. (1995). DNA (250 ng) was co-digested with MseI and EcoRI at 37°C for 2 h and oligomer adapters ligated to DNA fragments at 37°C for 3 h in 40 μL of digestion-ligation buffer. Preselective amplification was performed with 5 μL of digestion-ligation reaction in 50 μL of polymerase chain reaction (PCR) buffer containing nonselective primers MseI+0 and EcoRI+0 (20 cycles of 30 s at 94°C, 60 s at 56°C, and 60 s at 72°C). Selective amplification was performed with 5 μL of 1:30 diluted preselective amplification reaction in 20 μL of PCR buffer containing primers MseI + A and 33P-labelled EcoRI + AGG (one cycle of 30 s at 94°C, 30 s at 65°C, and 60 s at 72°C; 12 cycles of 65°C with annealing temperature lowered by 0.7°C during each cycle; and 23 cycles of 30 s at 94°C, 30 s at 56°C, and 60 s at 72°C).
Amplified DNA fragments were separated on a 6% polyacrylamide gel electrophoresed at 50 W for 2.5 h on an AFLexpress automatic sequencer (Amersham Pharmacia Biotech, Roosendaal, the Netherlands) flanked by a 30–330 plus 1668 bp AFLP DNA ladder. Autoradiographs were obtained by exposing Kodak BioMax MR film (Eastman Kodak Co., Rochester, NY, USA) to dried gels. All AFLP bands of medium to dark intensity were scored manually from the autoradiographs. Fragment sizes were inferred using Gene Profiler Eval. 4.03 (Scanalytics, Rockville, MD, USA). A common set of four reference strains were included on each gel to maintain consistency of scoring across gels. Identical profiles were obtained from different DNA preparations of the same isolates, confirming the reproducibility of the AFLP fingerprints.
AFLP bands were scored as dominant markers (present/absent). The binary data matrix was analyzed using NTSYSpc 2.11X (Exeter Software, Setauket, NY, USA). Haplotypes were determined by calculating the Dice coefficient of genetic similarity in the SIMQUAL module and constructing an unweighted pair-group with arithmetic averages (UPGMA) dendrogram in the SAHN module. Bootstrap values (10 000 replicates) for each branch (%) of the dendrogram were calculated using Winboot (International Rice Research Institute, Manila, Philippines).
Sequence analysis
Amplification and sequencing primers are listed in Table 2. Portions of the translation elongation factor-1α (EF-1α) gene, the mitochondrial small subunit (mtSSU) rDNA, the nitrate reductase (NIR) gene, and the phosphate permease (PP) gene were amplified and sequenced from representative isolates (Table 3). The genes were amplified in 50 μL reaction mixtures containing 100 ng template DNA, 1.5 mm MgCl, 2 mm dNTPs, 10 pm primer, and 2 U Amplitaq DNA Polymerase (Applied Biosystems, Foster City, CA, USA) in 1×GeneAmp buffer (Applied Biosystems). PCR amplifications were performed in a Hybaid Express cycler (Thermo, San Diego, CA, USA) with the following program: initial denaturing (2 min at 95°C), 35 cycles of denaturing (30 s at 94°C), primer annealing, primer extension (45 s at 72°C), and final extension (5 min at 72°C). PCR products were purified using Amicon Montage PCR clean-up columns (Millipore, Bedford, MA, USA) and re-suspended in 100 μL of 10 mm TRIS.
Table 2.
Primers used in this study
| Locus | Primer sequence (5′ to 3′) | Length | Tm (°C) | References | Use |
|---|---|---|---|---|---|
| Translation elongation factor-1α (EF-1α) | |||||
| EF-1 | ATG GGT AAG GAA GAC AAG AC | 20 | 50 | O'Donnell et al. 1998b | Amp.; Seq. |
| EF-2 | GGA AGT ACC AGT GAT CAT GTT | 21 | 50 | O'Donnell et al. 1998b | Seq. |
| Mitochondrial small subunit (mtSSU) | |||||
| MS1 | CAG CAG TCA AGA ATA TTA GTC AAT G | 25 | 50 | White et al. 1990 | Amp.; Seq. |
| MS2 | GCG GAT TAT CGA ATT AAA TAA C | 22 | 55 | White et al. 1990 | Amp.; Seq. |
| Nitrate reductase (NIR) | |||||
| NIR 1F | CCG CGG GAT CAG ACC AGA GCC C | 22 | 60 | Skovgaard et al. 2001 | Amp.; Seq. |
| NIR 2R | TTT GGA GGT AGA GGA TAA GGC | 21 | 60 | Skovgaard et al. 2001 | Amp.; Seq. |
| Phosphate permease (PP) | |||||
| PHO1 | ATC TTC TGG CGT GTT ATC ATG | 21 | 50 | O'Donnell et al. 2000 | Amp.; Seq. |
| PHO3 | TTC CAG CAC TAC AGC AAG TGG | 21 | 65 | This study | Seq. |
| PHO4 | GTG CTG GAA GAA GTC TCT CC | 20 | 55 | O'Donnell et al. 2000 | Seq. |
| PHO6 | GAT GTG GTT GTA AGC AAA GCC C | 22 | 50 | O'Donnell et al. 2000 | Amp.; Seq. |
Tm, annealing temperature; Amp., amplification; Seq., sequencing.
Table 3.
Representative isolates from which translation elongation factor (EF-1α), mitochondrial small subunit (mtSSU) rDNA, nitrate reductase (NIR), and phosphate permease (PP) sequences were generated in this study to assess the phylogenetic relationships between Australian Fusarium oxysporum and F. oxysporum f. sp vasinfection (Fov) race 1–8 and between Australian F. oxysporum and those found elsewhere in the world
| Accession identifier | Pathogenicity on Gossypium hirsutum | Lineage | Subgroup/haplotype* | Origin, race or VCG group | EF-1α GenBank acc no. | mtSSU GenBank acc no. | NIR GenBank acc no. | PP GenBank acc no. |
|---|---|---|---|---|---|---|---|---|
| Accessions from Boggabilla site | ||||||||
| 7080 | Pathogenic | A | A-I/A02 | Field soil | DQ435339 | DQ435357 | –† | – |
| 251104 | Pathogenic | A | A-I/A05 | Diseased plant | DQ435340 | DQ435358 | – | – |
| 241117 | Pathogenic | A | A-II/A14 | Diseased plant | DQ435341 | DQ435359 | – | – |
| 7110 | Pathogenic | A | A-II/A16 | Field soil | DQ435342 | DQ435360 | – | – |
| 7094 | Nonpathogenic | A | A-I/A06 | Field soil | DQ435343 | DQ435361 | – | – |
| 7135 | Nonpathogenic | A | A-I/A08 | Field soil | DQ435344 | DQ435362 | – | – |
| 6521 | Nonpathogenic | A | A-I/A11 | Refuge soil | DQ435345 | DQ435363 | – | – |
| 7099 | Nonpathogenic | A | A-I/A12 | Field soil | DQ435436 | DQ435364 | – | – |
| 6543 | Nonpathogenic | A | A-I/A13 | Refuge soil | DQ435347 | DQ435365 | – | – |
| 7081 | Nonpathogenic | A | A-II/A19 | Field soil | DQ435348 | DQ435366 | – | – |
| 7108 | Nonpathogenic | B | /B26 | Field soil | DQ435349 | DQ435367 | – | – |
| 7070 | Nonpathogenic | E | /E32 | Field soil | DQ435350 | DQ435368 | – | – |
| Accessions from soil associated with wild Gossypium | ||||||||
| 2613 | Slightly pathogenic | A | A-I/A14 | Wild Gossypium soil | DQ435351 | DQ435369 | EU246622 | EU246656 |
| 3545 | Nonpathogenic | A | – | Wild Gossypium soil | EU246540 | EU246587 | EU246623 | EU246657 |
| 3546 | Slightly pathogenic | A | – | Wild Gossypium soil | EU246541 | EU246588 | EU246624 | EU246658 |
| 3547 | Slightly pathogenic | A | – | Wild Gossypium soil | EU246542 | EU246589 | EU246625 | EU246659 |
| 3549 | Nonpathogenic | A | A-I/A15 | Wild Gossypium soil | DQ435352 | DQ435370 | EU246626 | EU246660 |
| 3556 | Slightly pathogenic | A | – | Wild Gossypium soil | EU246543 | EU246590 | EU246627 | EU246661 |
| 3608 | Nonpathogenic | A | – | Wild Gossypium soil | EU246544 | EU246591 | EU246628 | EU246662 |
| 6510 | Nonpathogenic | A | – | Wild Gossypium soil | EU246545 | EU246592 | EU246629 | EU246663 |
| 6632 | Nonpathogenic | A | – | Wild Gossypium soil | EU246546 | EU246593 | EU246630 | EU246664 |
| 1517 | Slightly pathogenic | B | – | Wild Gossypium soil | EU246586 | EU246619 | – | – |
| 1537 | Nonpathogenic | B | – | Wild Gossypium soil | EU246585 | EU246620 | – | – |
| 2631 | Slightly pathogenic | E | – | Wild Gossypium soil | EU246562 | EU246597 | EU246634 | EU246668 |
| 3506 | Slightly pathogenic | E | – | Wild Gossypium soil | EU246563 | EU246598 | EU246635 | EU246669 |
| 3522 | Nonpathogenic | E | – | Wild Gossypium soil | EU246564 | EU246599 | EU246636 | EU246670 |
| 3544 | Nonpathogenic | E | – | Wild Gossypium soil | EU246565 | EU246621 | – | – |
| 3552 | Slightly pathogenic | E | – | Wild Gossypium soil | EU246566 | EU246600 | EU246637 | EU246671 |
| 4511 | Slightly pathogenic | E | – | Wild Gossypium soil | EU246567 | EU246601 | EU246638 | EU246672 |
| 4590 | Slightly pathogenic | E | – | Wild Gossypium soil | EU246568 | EU246602 | EU246639 | EU246673 |
| 6519 | Nonpathogenic | E | – | Wild Gossypium soil | EU246569 | EU246603 | EU246640 | EU246674 |
| Reference accessions obtained from public collections | ||||||||
| SC1‡ | Fov | – | – | Race 1 | EU246574 | EU246608 | EU246645 | EU246679 |
| IMI-141148‡ | Fov | – | – | Race 2 | EU246571 | EU246605 | EU246642 | EU246676 |
| IMI-338122‡ | Fov | – | – | Race 3 | EU246573 | EU246607 | EU246644 | EU246678 |
| IMI-141112‡ | Fov | – | – | Race 4 | EU246570 | EU246604 | EU246641 | EU246675 |
| IMI-325576‡ | Fov | – | – | Race 5 | EU246572 | EU246606 | EU246643 | EU246677 |
| ATCC-16611‡ | Fov | – | – | Race 6 | EU246549 | EU246596 | EU246633 | EU246667 |
| Ag6‡ | Fov | – | – | Race 7 | EU246547 | EU246594 | EU246631 | EU246665 |
| Ag85‡ | Fov | – | – | Race 8 | EU246548 | EU246595 | EU246632 | EU246666 |
| 24500§ | Fov | – | – | VCG 01111 | EU246575 | EU246609 | EU246646 | EU246680 |
| 24595§ | Fov | – | – | VCG 01111 | EU246576 | EU246610 | EU246647 | EU246681 |
| 041101 | Fov | – | – | VCG 01111 | EU246578 | EU246611 | EU246648 | EU246682 |
| 051101 | Fov | – | – | VCG 01111 | EU246579 | EU246612 | EU246649 | EU246683 |
| X1§ | Fov | – | – | VCG 01111 | EU246577 | EU246613 | EU246650 | EU246684 |
| 24492§ | Fov | – | – | VCG 01112 | EU246584 | EU246618 | EU246655 | EU246689 |
| 24597§ | Fov | – | – | VCG 01112 | EU246580 | EU246614 | EU246651 | EU246685 |
| 24598§ | Fov | – | – | VCG 01112 | EU246581 | EU246615 | EU246652 | EU246686 |
| 24646§ | Fov | – | – | VCG 01112 | EU246582 | EU246616 | EU246653 | EU246687 |
| B/96/02§ | Fov | – | – | VCG 01112 | EU246583 | EU246617 | EU246654 | EU246688 |
Identified based on results illustrated in Fig. 6.
Data not available.
Provided in the form of DNA by Linda Smith (Queensland Department of Primary Industries, Indooroopilly, Australia).
Provided in culture by Natalie Moore, Linda Smith, and Wayne O'Neil (Queensland Department of Primary Industries, Indooroopilly, Australia).
Sequencing reactions were conducted on the purified PCR products with 3.2 pm of the forward or reverse primer using the fluorescent-labeled BigDye kits v3.1 (Perkin-Elmer, Boston, MA, USA) in a Hybaid Express cycler (Thermo) with the program recommended by the manufacturer. Products were cleaned up by isopropanol precipitation and run on an ABI PRISM Genetic Analyzer capillary sequencer (Applied Biosystems).
Forward and reverse sequences were assembled, edited using Sequencher 4.2 (Gene Codes, Ann Arbor, MI, USA), and deposited in GenBank (Table 3). Alignments were conducted using ClustalW as implemented in BioEdit 7.0.5.2 (Hall 1999). In addition to the sequences generated in this study, representatives of F. oxysporum f. sp. vasinfectum race 1–8 and representative taxa from the order Hypocreales were downloaded from GenBank to augment the alignments (Table 4). Three sequence alignments were constructed: (i) concatenated EF-1α and mtSSU sequences from representative strains of Fov from Boggabilla and native F. oxysporum from soils associated with wild Gossypium populations to explore the genetic relationships between pathogenic and nonpathogenic (against G. hirsutum cotton) Australian strains (deposited in TreeBASE under the accession numbers SN2747-10816); (ii) concatenated EF-1α, mtSSU, NIR, and PP sequences from pathogenic and nonpathogenic Australian isolates and representatives of Fov races 1–8 to determine the genetic relationships between Australian F. oxysporum strains and Fov occurring elsewhere in the world (deposited in TreeBASE under the accession numbers SN3665-16634); and (iii) EF-1α sequences from all lineages of Australian F. oxysporum identified by the AFLP analyses and representatives of other key Fusarium lineages to assess the phylogenetic relationships of the Australian F. oxysporum to other Fusarium species and taxa (deposited in TreeBASE under the accession numbers SN3665-16635).
Table 4.
Translation elongation factor (EF-1α), mitochondrial small subunit (mtSSU) rDNA, nitrate reductase (NIR), and phosphate permease (PP) sequences from representative Fusarium, Gibberella, Nectria, and Neocosmospora species that were used to assess the relationships of Australian F. oxysporum to other major phylogenetic groups within the order Hypocreales
| Accession identifier | Genus | Species | EF-1α GenBank acc no. | mtSSU GenBank acc no. | NIR GenBank acc no. | PP GenBank acc no. |
|---|---|---|---|---|---|---|
| NRRL25300 | Fusarium | begoniae | AF160293 | –* | – | – |
| NRRL31238 | Fusarium | brasilicum | AY452963 | – | – | – |
| NRRL31281 | Fusarium | brasilicum | AY452964 | – | – | – |
| NRRL22678 | Fusarium | brasiliense | AY320144 | – | – | – |
| NRRL22743 | Fusarium | brasiliense | AY320145 | – | – | – |
| NRRL13618 | Fusarium | bulbicola | AF160294 | – | – | – |
| NRRL13721 | Fusarium | cerealis | AF212464 | – | – | – |
| NRRL25491 | Fusarium | cerealis | AF212465 | – | – | – |
| NRRL28387 | Fusarium | commune | AF246832 | – | – | – |
| NRRL26434 | Fusarium | concentricum | AF333933 | – | – | – |
| NRRL31171 | Fusarium | cortaderiae | AY452961 | – | – | – |
| NRRL31205 | Fusarium | cortaderiae | AY452960 | – | – | – |
| NRRL25475 | Fusarium | culmorum | AF212463 | – | – | – |
| NRRL3288 | Fusarium | culmorum | AF212462 | – | – | – |
| NRRL22275 | Fusarium | cuneirostrum | AY320158 | – | – | – |
| NRRL31104 | Fusarium | cuneirostrum | AY320159 | – | – | – |
| NRRL31044 | Fusarium | foetens | AY320072 | – | – | – |
| NRRL31045 | Fusarium | foetens | AY320073 | – | – | – |
| NRRL28854 | Fusarium | fractiflexum | AF333932 | – | – | – |
| NRRL22945 | Fusarium | guttiforme | AF160297 | – | – | – |
| NRRL29642 | Fusarium | hostae | AF324322 | – | – | – |
| NRRL29643 | Fusarium | hostae | AF324323 | – | – | – |
| NRRL25200 | Fusarium | lactis | AF160272 | – | – | – |
| VI01268 | Fusarium | langsethiae | AJ420822 | – | – | – |
| VI01271 | Fusarium | langsethiae | AJ420823 | – | – | – |
| NRRL26231 | Fusarium | miscanthi | AF324331 | – | – | – |
| NRRL26239 | Fusarium | miscanthi | AF324332 | – | – | – |
| NRRL13604 | Fusarium | napiforme | AF160266 | – | – | – |
| NRRL25179 | Fusarium | nisikadoi | AF324329 | – | – | – |
| NRRL25183 | Fusarium | nisikadoi | AF324330 | – | – | – |
| BBA65634 | Fusarium | oxysporum f. sp. vasinfectum (race 1) | AF362145 | AF362178 | AF362145 | AF362178 |
| BBA64495 | Fusarium | oxysporum f. sp. vasinfectum (race 2) | AF362144 | AF362177 | AF362144 | AF362177 |
| BBA65633 | Fusarium | oxysporum f. sp. vasinfectum (race 2) | AF362146 | AF362179 | AF362146 | AF362179 |
| BBA65635 | Fusarium | oxysporum f. sp. vasinfectum (race 2) | AF362147 | AF362180 | AF362147 | AF362180 |
| BBA65636 | Fusarium | oxysporum f. sp. vasinfectum (race 2) | AF362148 | AF362181 | AF362148 | AF362181 |
| BBA65653 | Fusarium | oxysporum f. sp. vasinfectum (race 2) | AF362141 | AF362174 | AF362141 | AF362174 |
| BBA65655 | Fusarium | oxysporum f. sp. vasinfectum (race 2) | AF362149 | AF362182 | AF362149 | AF362182 |
| BBA66844 | Fusarium | oxysporum f. sp. vasinfectum (race 2) | AF362150 | AF362183 | AF362150 | AF362183 |
| BBA69405 | Fusarium | oxysporum f. sp. vasinfectum (race 2) | AF362151 | AF362184 | AF362151 | AF362184 |
| BBA62374 | Fusarium | oxysporum f. sp. vasinfectum (race 3) | AF362142 | AF362175 | AF362142 | AF362175 |
| BBA62375 | Fusarium | oxysporum f. sp. vasinfectum (race 3) | AF362143 | AF362176 | AF362143 | AF362176 |
| BBA64496 | Fusarium | oxysporum f. sp. vasinfectum (race 3) | AF362159 | AF362192 | AF362159 | AF362192 |
| BBA66845 | Fusarium | oxysporum f. sp. vasinfectum (race 3) | AF362153 | AF362186 | AF362153 | AF362186 |
| BBA67521 | Fusarium | oxysporum f. sp. vasinfectum (race 3) | AF362152 | AF362185 | AF362152 | AF362185 |
| BBA69712 | Fusarium | oxysporum f. sp. vasinfectum (race 3) | AF362162 | AF362195 | AF362162 | AF362195 |
| BBA66846 | Fusarium | oxysporum f. sp. vasinfectum (race 4) | AF362164 | AF362197 | AF362164 | AF362197 |
| BBA69518 | Fusarium | oxysporum f. sp. vasinfectum (race 4) | AF362160 | AF362193 | AF362160 | AF362193 |
| BBA69519 | Fusarium | oxysporum f. sp. vasinfectum (race 4) | AF362157 | AF362190 | AF362157 | AF362190 |
| BBA69520 | Fusarium | oxysporum f. sp. vasinfectum (race 4) | AF362140 | AF362173 | AF362140 | AF362173 |
| BBA69521 | Fusarium | oxysporum f. sp. vasinfectum (race 4) | AF362139 | AF362172 | AF362139 | AF362172 |
| BBA65650 | Fusarium | oxysporum f. sp. vasinfectum (race 5) | AF362154 | AF362187 | AF362154 | AF362187 |
| BBA65654 | Fusarium | oxysporum f. sp. vasinfectum (race 5) | AF362155 | AF362188 | AF362155 | AF362188 |
| BBA66847 | Fusarium | oxysporum f. sp. vasinfectum (race 6) | AF362158 | AF362191 | AF362158 | AF362191 |
| BBA69716 | Fusarium | oxysporum f. sp. vasinfectum (race 7) | AF362163 | AF362196 | AF362163 | AF362196 |
| BBA69050 | Fusarium | oxysporum f. sp. vasinfectum (race 7) | AF362156 | AF362189 | AF362156 | AF362189 |
| BBA69711 | Fusarium | oxysporum f. sp. vasinfectum (race 8) | AF362161 | AF362194 | AF362161 | AF362194 |
| NRRL22276 | Fusarium | phaseoli | AY220186 | – | – | – |
| NRRL31156 | Fusarium | phaseoli | AY220187 | – | – | – |
| NRRL31071 | Fusarium | proliferatum | AF291058 | – | – | – |
| NRRL22946 | Fusarium | pseudocircinatum | AF160271 | – | – | – |
| NRRL25208 | Fusarium | ramigenum | AF160267 | – | – | – |
| NRRL25600 | Fusarium | redolens | AF324294 | – | – | – |
| NRRL28181 | Fusarium | redolens | AF077391 | – | – | – |
| NRRL22400 | Fusarium | solani f.sp. batas | AF178343 | – | – | – |
| NRRL22402 | Fusarium | solani f.sp. batas | AF178344 | – | – | – |
| VI01313 | Fusarium | sporotrichioides | AJ420818 | – | – | – |
| VI01319 | Fusarium | sporotrichioides | AJ420819 | – | – | – |
| NRRL13613 | Fusarium | succisae | AF160291 | – | – | – |
| NRRL31085 | Fusarium | tucumaniae | AY220170 | – | – | – |
| NRRL31086 | Fusarium | tucumaniae | AY220171 | – | – | – |
| NRRL22292 | Fusarium | virguliforme | AY220188 | – | – | – |
| NRRL22489 | Fusarium | virguliforme | AY220189 | – | – | – |
| NRRL26432 | Gibberella | circinata | AF333929 | – | – | – |
| NRRL28894 | Gibberella | moniliformis | AF273313 | – | – | – |
| NRRL29169 | Gibberella | zeae | AF212461 | – | – | – |
| NRRL34079 | Gibberella | zeae | AY452958 | – | – | – |
| NRRL22141 | Nectria | haematococca | AF178329 | – | – | – |
| NRRL22161 | Nectria | haematococca | AF178330 | – | – | – |
| NRRL22436 | Neocosmospora | africana | AF178348 | – | – | – |
| NRRL22468 | Neocosmospora | ornamentata | AF178349 | – | – | – |
| NRRL22166 | Neocosmospora | vasinfecta | AF178350 | – | – | – |
Data not available.
Parsimony optimized topologies, partition homogeneity estimates, and bootstrap values were generated using PAUP 4.0 beta 10 (Sinauer Associates, Sunderland, MA, USA). Unweighted maximum parsimony was conducted using the heuristic search option and 100 random addition sequences with the tree-bisection-reconnection branch swapping and the MULTREES option on. Bayesian inference was used to estimate posterior probabilities for consensus nodes using MRBAYES 3.1 (Ronquist and Huelsenbeck 2003) and the most appropriate models of sequence evolution for the Bayesian analysis were identified using Modeltest 3.7 (Posada and Crandall 1998). Trees were visualized using TreeView 1.6.6 (Page 1996).
VCG tests
The vegetative compatibility of native F. oxysporum isolates derived from soils associated with wild Gossypium and reference Fov strains were tested using the method described by Puhalla (1985). For each strain, three nitrate nonutilizing mutants (nit 1, nit 3, and Nit M) were generated on a minimal medium amended with 1.5–4.0% (w/v) of potassium chlorate. Pairing tests were performed in 96 cell plates by growing different mutants of two isolates at 25°C for 2 weeks in a minimal medium containing sodium nitrate as the sole nitrogen source. Heterokaryon formation was identified by wild-type growth.
Results
Fungal isolation
A total of 856 F. oxysporum isolates were recovered, including 562 isolates from soils associated with wild Gossypium, 35 from uncultivated refuge soil, 178 from cultivated field soils, and 81 from diseased cotton plants (Table 5). The incidence of F. oxysporum varied among species and regions, occurring in 91% of soils associated with G. sturtianum populations, but in only 48–62% of populations of the other three species. Among the four regions, F. oxysporum occurred at a high frequency in the Longreach-Theodore (92%) and Leigh Creek-Arkaroola (85%) regions, but at a lower frequency in the Alice Spring-Tennant Creek (59%) and Mount Isa regions (57%) (Table 1). Fusarium oxysporum was isolated from all uncultivated and cultivated soils collected from the Boggabilla region, and Fov was isolated from all diseased G. hirsutum plants.
Table 5.
Number of isolates of Fusarium oxysporum that are nonpathogenic on cotton (NP) and isolates of F. oxysporum f. sp. vasinfectum (Fov) recovered from Gossypium soil (summarized by Gossypium species and geographic regions respectively), uncultivated refuge soil, cultivated field soil, and diseased cotton plants, by lineage
| Lineage | ||||||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | Others | Total | ||
| Source | Path* | N† (%)‡ | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) |
| Gossypium soil by Gossypium species | ||||||||
| G. australe | NP | 0 (0) | 82 (59) | 36 (26) | 0 (0) | 2 (1) | 5 (4) | 125 (89) |
| Fov§ | 0 (0) | 10 (7) | 2 (1) | 0 (0) | 1 (1) | 2 (1) | 15 (11) | |
| G. bickii | NP | 0 (0) | 36 (73) | 0 (0) | 0 (0) | 0 (0) | 6 (12) | 42 (86) |
| Fov | 0 (0) | 6 (12) | 0 (0) | 0 (0) | 0 (0) | 1 (2) | 7 (14) | |
| G. nelsonii | NP | 0 (0) | 47 (77) | 5 (8) | 2 (3) | 0 (0) | 1 (2) | 55 (90) |
| Fov | 0 (0) | 6 (10) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 6 (10) | |
| G. sturtianum | NP | 18 (6) | 144 (46) | 52 (17) | 3 (1) | 25 (8) | 14 (4) | 256 (82) |
| Fov | 14 (4) | 19 (6) | 0 (0) | 2 (1) | 17 (5) | 4 (1) | 56 (18) | |
| Gossypium soil by geographic regions | ||||||||
| Mount Isa (QLD) | NP | 0 (0) | 18 (16) | 87 (76) | 2 (2) | 0 (0) | 2 (2) | 109 (95) |
| Fov | 0 (0) | 4 (3) | 2 (2) | 0 (0) | 0 (0) | 0 (0) | 6 (5) | |
| Longreach (QLD)¶ | NP | 1 (1) | 102 (82) | 0 (0) | 0 (0) | 1 (1) | 4 (3) | 108 (86) |
| Fov | 1 (1) | 11 (9) | 0 (0) | 0 (0) | 4 (3) | 1 (1) | 17 (14) | |
| Alice Springs (NT)** | NP | 1 (1) | 146 (74) | 6 (3) | 3 (2) | 4 (2) | 11 (6) | 171 (86) |
| Fov | 1 (1) | 20 (10) | 0 (0) | 2 (1) | 1 (1) | 3 (2) | 27 (14) | |
| Leigh Creek (SA)†† | NP | 16 (13) | 43 (35) | 0 (0) | 0 (0) | 22 (18) | 9 (7) | 90 (73) |
| Fov | 12 (10) | 6 (5) | 0 (0) | 0 (0) | 13 (10) | 3 (2) | 34 (27) | |
| Gossypium soil | NP | 18 (3) | 309 (55) | 93 (17) | 5 (1) | 27 (5) | 26 (5) | 478 (85) |
| Fov | 14 (2) | 41 (7) | 2 (0) | 2 (0) | 18 (3) | 7 (1) | 84 (15) | |
| Refuge soil | NP | 2 (6) | 32 (91) | 0 (0) | 0 (0) | 1 (3) | 0 (0) | 35 (100) |
| Fov | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Field soil | NP | 11 (6) | 4 (2) | 0 (0) | 0 (0) | 145 (81) | 0 (0) | 160 (90) |
| Fov | 18 (10) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 18 (10) | |
| Diseased plants | NP | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Fov | 81 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 81 (100) | |
Pathogenicity of the isolates against cotton (G. hirsutum).
Number of isolates.
Percentage of the isolates in the total recovered.
Weakly pathogenic wild Fov for those isolates from Gossypium soils.
Longreach-Theodore (QLD).
Alice Springs-Tennant Creek (NT).
Leigh Creek-Arkaroola (SA).
Pathogenicity screening tests
Fifteen percent of the 562 F. oxysporum isolates from soils associated with native Gossypium, were weakly pathogenic (i.e. causing mild stunting, foliar necrosis, and vascular discoloration) on Siokra 1–4, one of the most susceptible Australia cotton cultivars, with a mean disease severity of 0.3 (range: 0.1–0.6). This group was therefore putatively designated as wild Fov (Table 5). In contrast to the Fov found in cotton fields, no isolate of wild Fov associated with wild Gossypium soils was able to kill inoculated plants during the 6-week experimental period.
The incidence of wild Fov among F. oxysporum isolates varied by Gossypium species as well as geographic region. The greatest incidence occurred in isolates derived from soils associated with G. sturtianum (18%), with lower numbers among isolates from the other three species (Table 5). The incidence of wild Fov also appeared to vary geographic, ranging from 27% of the Leigh Creek-Arkaroola region isolates to only 5% of those from the Mount Isa region (Table 5).
Eighteen (10%) of the 178 F. oxysporum isolates recovered from cultivated field soils were Fov causing severe wilt symptoms in both trials (mean disease severity = 2.5; range = 1.8–3.1). However, none of the isolates from the uncultivated soil was pathogenic on cotton (Table 5). All isolates from diseased cotton plants were confirmed to be Fov as they consistently caused severe disease symptoms in both pathogenicity screening trials.
Virulence comparison tests
Wild F. oxysporum f. sp. vasinfectum (strains 2613 and 3556 from soils associated with G. sturtianum) was less aggressive on cotton but similar, or even more aggressive, on G. sturtianum relative to the performance of the reference Fov strains (Fig. 2). The two wild Fov strains caused only slight disease symptoms on cotton cultivar Sicot 189 with severity ranging from 0.1 to 0.5, whereas the two reference Fov strains (derived from diseased cotton plants) caused significantly more severe disease symptoms (range: 1.3–2.9). Plants of G. sturtianum Gos-5250 were susceptible to both the wild and reference Fov strains (severity range: 1.3–3.1). While no significant difference in disease severity was found between wild Fov strain 3556 and the two reference Fov strains on Gos-5250, wild Fov strain 2613 caused significantly more severe disease symptoms (Fig. 2).
Figure 2.
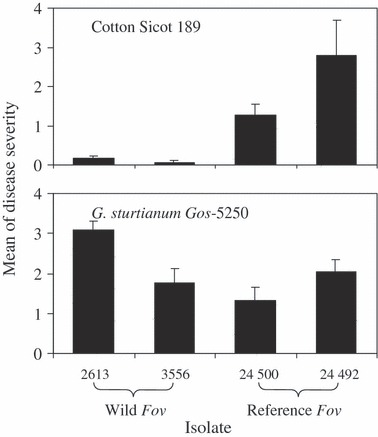
Mean disease severity on plants of cotton (Sicot 189) and Gossypium sturtianum (Gos 5250) caused by wild Fusarium oxysporum f. sp. vasinfectum (Fov) from soil associated with wild Gossypium (strains 2613 and 3556) and the Fov found in cotton fields (VCG 01111 strain 24500 and VCG 011112 strain 24492). Bars on top of columns represent standard deviations.
AFLP analysis
Of the 562 isolates of native F. oxysporum from soil associated with wild Gossypium, 94% (529) were grouped into five genetic lineages designated A, B, C, D, and E (Table 5). The lineage groupings were supported by the results of both an UPGMA (similarities between any two lineages <50%), and a bootstrap analysis, in which the bootstrap values based on data from four representatives per lineage ranged from 95 to 100 (Fig. 3).
Figure 3.
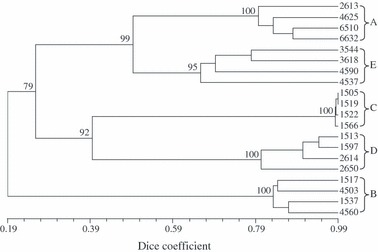
Unweighted pair-group with arithmetic average dendrogram constructed from AFLP fingerprints of four representative strains of each of the five genetic lineages (A, B, C, D and E) of Australian Fusarium oxysporum identified in this study. A total of 137 polymorphic bands revealed by the primer combination of EcoRI + AC and MseI + A were used. Numbers at the nodes of major clusters represent bootstrap values (%) generated by 1000 replicates.
The distribution of lineages in isolates from soil associated with wild Gossypium varied among species and region (Table 5). Lineage B predominated in four of the five regions irrespective of host species while lineages A and E were similarly distributed but were concentrated in the Leigh Creek-Arkaroola region. In contrast, lineage C was restricted to the Mount Isa region where it predominated; and only lineage B was found in association with G. bickii populations (Table 5).
The incidence of wild Fov in soils associated with wild Gossypium also varied among the five lineages, ranging from c. 40% in lineages A and E, down to 11% and 2% in lineages B and C respectively. The incidence of wild Fov in lineage D (29%) is based on the occurrence of only two individuals in a sample size of seven (Table 5).
Lineages A, B, and E were also found in Boggabilla soil and plant samples (Table 5; Fig. 4). Ninety-five genetically distinct haplotypes clustering into three well-supported lineages (bootstrap values 100%) were identified among the 294 isolates from this area. Nineteen, 26, and 50 haplotypes were detected in lineages A, B and E, respectively. The level of genetic similarity among haplotypes within lineages was relatively high (72–75%), while genetic similarities between isolates from different lineages was considerably lower – 50% between lineages A and E, and only 13% between isolates in lineage B and those in lineages A or E. Lineage A could be further divided into two subgroups (A-I and A-II), but no clear subdivision was distinguishable in the other lineages (Fig. 4).
Figure 4.
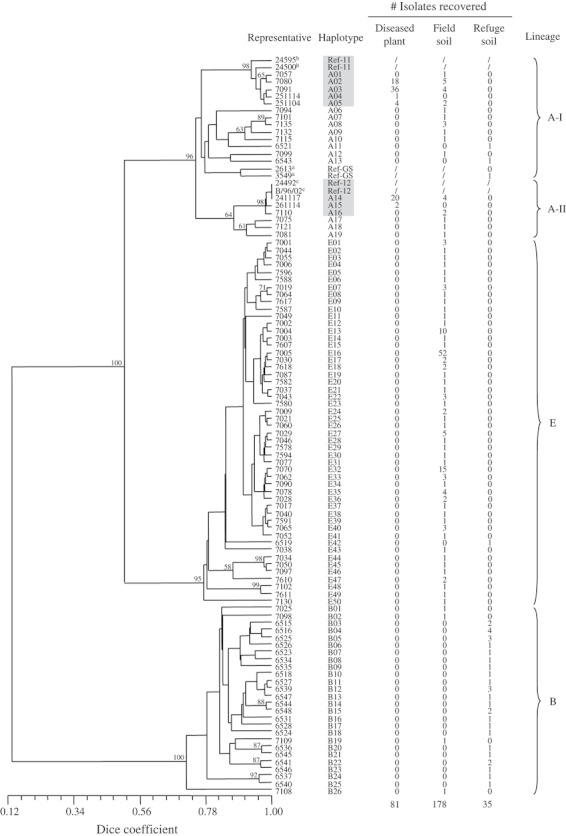
A UPGMA dendrogram illustrating genetic relationships among 95 haplotypes of nonpathogenic Fusarium oxysporum (against cotton) and Fov based on pairwise Dice estimates of genetic similarity revealed using AFLPs. Bootstrap values ≥58% (10 000 replicates) are shown above nodes. Haplotypes in shaded boxes are pathogenic, i.e. Fov. The number of isolates per haplotype from diseased plants, cultivated fields, and refuge soil are listed to the right of the dendrogram. Brackets at the extreme right of the figure denote the lineages and subgroups discussed in the text. Isolates numbers followed by a superscript ‘a’, ‘b’, and ‘c’ refer to representative strains from soil associated with wild Gossypium, VCG 01111 and VCG 01112 of the Australian Fov, respectively.
All pathogenic isolates (i.e. Fov) from Boggabilla belonged to lineage A, regardless of origin (Table 5; Fig. 4). They were distributed among eight haplotypes with five (A01-A05) in subgroup A-I and three (A14-A16) in subgroup A-II. Both reference Fov strains of VCG 01111 fell within subgroup A-I and both reference strains of VCG 01112 were placed in subgroup A-II. Of the eight Fov haplotypes, four (A02, A03, A05, A14) were recovered from both diseased plants and cultivated field soil, two (A04, A15) were found only in diseased plants, and two (A01, A16) only in the soil (Fig. 4). Thirteen nonpathogenic strains (fields: 11; refuge: 2) clustered with the pathogenic lineage A isolates, with eight in subgroup A-I and three in subgroup A-II. They were highly variable and represented a range of different haplotypes. These nonpathogenic lineage A isolates probably represented local Australian relatives of Fov (Fig. 4).
Ninety-three percent (182) of the nonpathogenic isolates from Boggabilla belonged to lineages B and E but the spatial distribution of these was almost mutually exclusive. Lineage E accounted for 82% of isolates from cultivated fields while 91% of isolates from the refuge soil were attributable to lineage B (Table 5). None of lineage A, B, or E nonpathogenic haplotypes were common to both the cultivated field and refuge soils (Fig. 4).
Sequence analysis
In the initial phylogenetic analysis concatenated sequences of two genes (EF-1α, mtSSU) from 18 isolates representing lineage A (12), B (3), and E (3) were used to explore relationships among the Australian Fov and nonpathogenic F. oxysporum (Fig. 5). The combined sequence alignment comprised 1404 base pairs (EF-1α: 738 bp; mtSSU: 666 bp), of which 66 were phylogenetically informative. No signal incongruence between the EF-1α and mtSSU genes was detected using the partition homogeneity test (P = 1.00).
Figure 5.
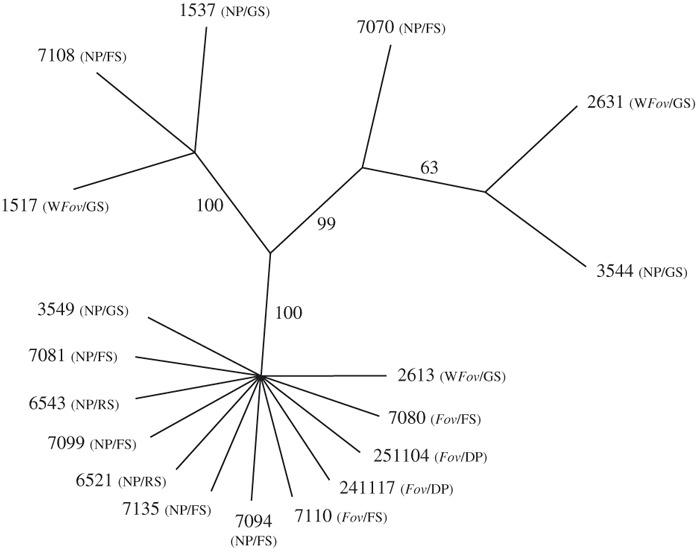
The single most parsimonious unrooted topology (length = 75 steps; consistency index = 0.9867; retention index = 0.9939) obtained from a heuristic parsimony optimized analysis of a concatenated matrix of the translation elongation factor-1α (EF-1α) gene and mitochondrial small subunit (mtSSU) rDNA sequences from four Fov, eight lineage A, three lineage B, and three lineage E isolates. Bootstrap values (10 000 replicates) are placed beside each branch of the typology. The pathogenicity of isolates on cotton (before slash; Fov = F. oxysporum f. sp. vasinfectum; WFov = wild F. oxysporum f. sp. vasinfectum; NP = nonpathogenic) and their origin (behind slash; GS = Gossypium soil; RS = uncultivated refuge soil; FS = cultivated field soil; DP = diseased plant) are given in brackets.
Unweighted maximum parsimony analysis yielded a single most parsimonious tree of 75 steps in length (consistency index = 0.987; retention index = 0.994; Fig. 5). The branching pattern of the tree was congruent with the UPGMA dendrograms of AFLP fingerprints (Figs 3 and 4), and further demonstrated that strains cluster based on genetic similarity rather than level of pathogenicity on cotton. Thus, all pathogenic and nonpathogenic isolates of lineage A were clustered in a well supported group (bootstrap value 100%), while lineage B and E strains occurred in two distinct and equally well supported clades that included nonpathogenic and weakly pathogenic wild Fov strains (Fig. 5).
In the second phylogenetic analysis, the relationships of the Australian Fov (including VCG 01111 and VCG 01112) and nonpathogenic lineage A isolates to representatives of Fov race 1–8 from other regions of the world was explored using concatenated sequences from four genes (EF-1α, mtSSU, NIR, PP). Data for races 1–8 was obtained from GenBank (Table 3). The concatenated alignment was 2399 bp in length, and four indels, encoded as binary characters, were appended to the end (EF-1α: 653 bp + two indels; mtSSU: 677 bp + one indel; NIR: 483 bp + one indel; PP: 586 bp). Unweighted maximum parsimony analysis yielded five equally parsimonious trees. There was no significant evidence of signal incongruity in the mtSSU sequence relative to the EF-1α, NIR, and PP sequences, and hence a parsimony optimized topology was derived from the unmodified alignment. To accommodate signal variation, the F81 substitution model was applied to the mtSSU partition, while the HKY model was applied to the EF-1α, NIR, and PP partitions in the Bayesian estimation of posterior probabilities (consistency index = 0.929; retention index = 0.992; Fig. 6). This topology demonstrates that the Australian Fov strains (VCG 01111 and VCG 01112) share a more recent common ancestor with both nonpathogenic and weakly pathogenic lineage A isolates of native F. oxysporum from soils associated with wild Gossypium than they do with Fov race 1–8 from overseas.
Figure 6.
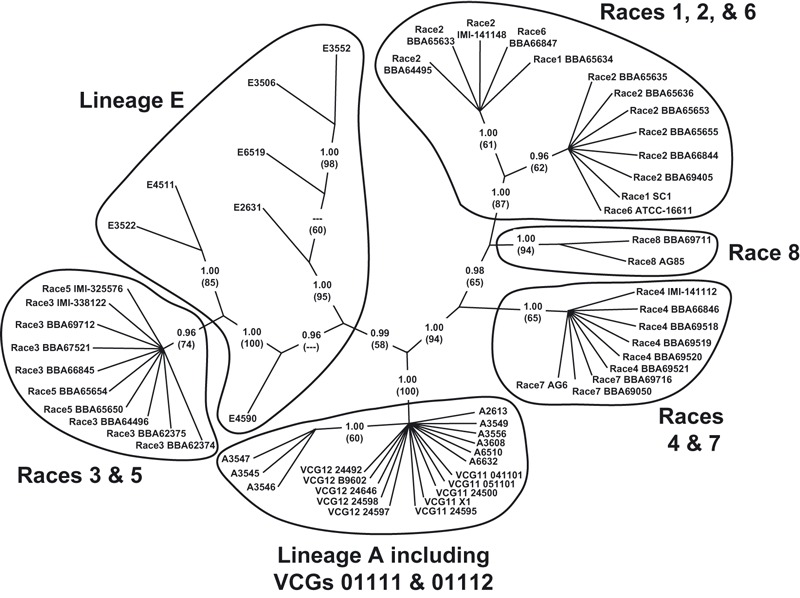
Consensus parsimony optimized phylogenetic tree based on concatenated alignment (2399 bp + four indels encoded as binary characters) of the translation elongation factor-1α (653 bp + two indels), mitochondrial small subunit rDNA (677 bp + one indel), nitrate reductase (483 bp + one indel), and phosphate permease (586 bp) gene sequences. This unrooted topology is the consensus of five equally parsimonious trees (consistency index = 0.9286; retention index = 0.9915). Posterior probabilities, estimated in a separate Bayesian analysis, are indicated for each node; bootstrap values (1000 replicates) are indicated parenthetically beneath each posterior probability.
In the final phylogenetic analysis, the relationships between characteristic Australian F. oxysporum-like isolates and representatives of key Fusarium lineages in the order Hypocreales were assessed using EF-1α sequences. This analysis was limited to a single gene to maximize the ability to incorporate a wider diversity of taxa. The alignment comprised 709 bp of EF-1α sequence appended by 51 indels encoded as binary characters for a composite length of 760 characters. The topology illustrated in Fig. 7 is an unrooted consensus of 24 equally parsimonious trees (consistency index = 0.642; retention index = 0.948). This topology (i) confirms the close phylogenetic relationships among the pathogenic (VCG 01111 and 01112) and nonpathogenic lineage A isolates evident in Figs 5 and 6; (ii) illustrates the sister relationships between the lineage A isolates and F. oxysporum f. sp. vasinfectum race 1–8 relative to F. foetens; (iii) reaffirms the close phylogenetic relationships among the Australian lineage E isolates and F. oxysporum f. sp. vasinfectum race 1–8 (see Fig. 6); (iv) suggests the lineage B is a component of the widespread F. fujikuroi complex; and (v) suggests that lineage C and D represent fungi that heretofore have not been sequenced possibly representing new taxa.
Figure 7.
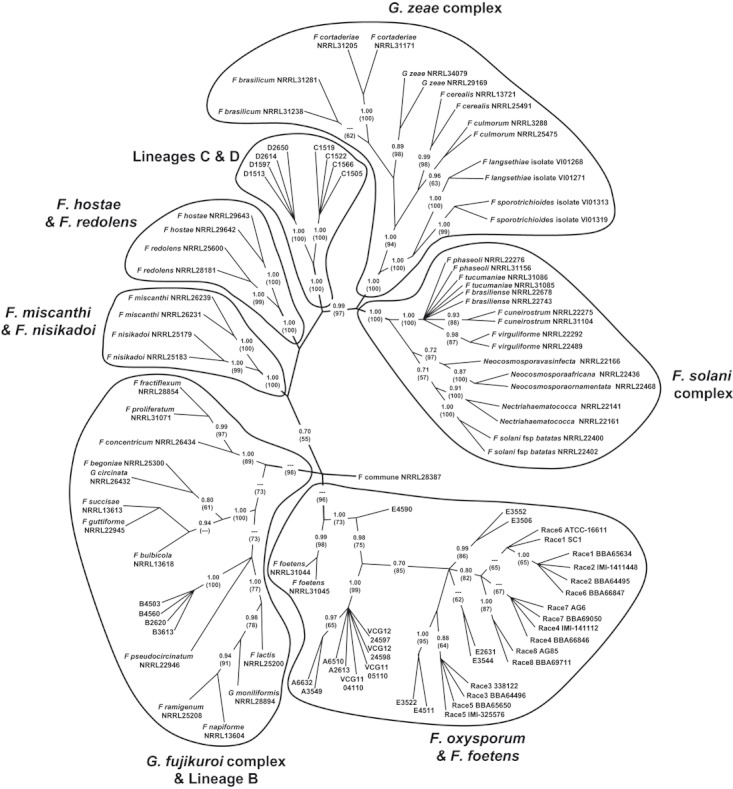
Consensus parsimony optimized phylogenetic tree based on the translation elongation factor-1α gene sequences (709 bp + 51 indels encoded as binary characters) from the Australian Fusarium oxysporum-like isolates and representatives of other key Fusarium lineages in the order Hypocreales. This unrooted topology is the consensus of 24 equally parsimonious trees (consistency index = 0.6416; retention index = 0.9480). Posterior probabilities, estimated in a separate Bayesian analysis, are indicated for each node; bootstrap values (1000 replicates) are indicated parenthetically beneath each posterior probability.
VCG tests
None of the 32 lineage A isolates derived from soil associated with wild Gossypium (Table 5) were compatible with either of the VCG associated with the four reference Australian Fov strains. Three successful pairings were observed among six isolates from Leigh Creek-Arkaroola, while the remaining isolates were incompatible with each other.
Discussion
Our results provide strong support for the hypothesis that VCG 01111 and VCG 01112 of Fov evolved locally in Australia (Davis et al. 1996). Not only are the Australian Fov strains morphologically and genetically distinct from the eight races of Fov found in other countries (Bayaa et al. 1995; Davis et al. 1996; Kim et al. 2005), but each VCG of the Australian Fov is grouped with a cluster of local nonpathogenic F. oxysporum strains in a single discrete lineage (lineage A; Fig. 4). This is further supported by multiple phylogenetic analyses that consistently place Australian Fov as sister to native lineage A strains rather than to representatives of the eight races of Fov that occur elsewhere in the world (Figs 5–7). That these putative precursors are endemic to Australia is well supported by their presence in both cultivated and uncultivated soils in cotton growing areas, and in a wide variety of soils associated with wild Gossypium located away from agricultural regions. The observation that some nonpathogenic lineage A strains are more related to the VCG 01111 strains than they are to the VCG 01112 strains, and the converse (Fig. 4), suggests that VCG 01111 and VCG 01112 arose independently.
The hypothesis that VCG 01111 and VCG 01112 of Fov evolved within Australia would be further strengthened by detection of their nonpathogenic progenitors, that is, native lineage A strains that are vegetatively compatible with the two known VCGs. However, the nonpathogenic lineage A strains found in soils associated with wild Gossypium in this study are highly unlikely to be direct progenitors of either VCG 01111 or VCG 01112 as none of them were vegetatively compatible. This is consistent with the observation of genetic similarity among the six compatible lineage A isolates. The minimum genetic similarity among these isolates is 94%, while the greatest genetic similarity between nonpathogenic isolates and pathogenic isolates obtained in this study was only 86% (Fig. 4).
The complexity of establishing the origins of pathogenic F. oxysporum is evident in a number of Fusarium wilt disease complexes (Gordon and Okamoto 1992a; Appel and Gordon 1994, 1996; Skovgaard et al. 2002). As is the case here, conclusions regarding the origin of newly emergent pathogens are often complicated by difficulty in discriminating between nonpathogenic progenitors and avirulent mutants of pathogenic strains. Some of the ambiguity could arise from different expectations regarding genetic relationships and vegetative compatibilities between pathogenic F. oxysporum that have arisen locally and co-occurring nonpathogenic populations. A reasonable expectation is that a pathogen may still be identical or similar to its ancestral strains, and consequently, most searches for the ancestors of pathogens focus on the same VCG (Gordon and Okamoto 1992b; Appel and Gordon 1994; Katan et al. 1994; Woudt et al. 1995). This is biologically realistic because individuals within a VCG are probably clonally derived, and genetic variation arises from mutation or other nonsexual means. Following this reasoning, F. oxysporum f. sp. vasinfectum and f. sp. lycopersici were regarded as exotic to Israel and California, respectively, because nonpathogenic forms that were vegetatively compatible with the pathogens were not observed in the soil community (Katan and Katan 1988; Elias et al. 1991). Conversely, the recovery of local, nonpathogenic F. oxysporum strains that were vegetatively compatible with races 1 and 2 of F. oxysporum f. sp. melonis in VCG 0134 in Maryland suggested a local origin for that pathogen (Appel and Gordon 1994).
However, while it is expected that recently emerged pathogens will be vegetatively compatible with their nonpathogenic progenitors, this expectation may never be realized when attempting to address older derivative–progenitor relationships. It cannot be assumed that the evolution of pathogenicity is temporally linked with the appearance of disease epidemics in an agricultural crop. For example, the ability of wild Fov strain 2613 (from soils associated with wild Gossypium) to cause mild Fusarium wilt symptoms on cotton suggests that some lineage A isolates could be characterized as ‘aggressive endophytes’, i.e. they can colonize the vasculature of cotton plants to the extent that some mild but typical disease symptoms are evident. If this is the case, then among a genetically diverse pool of native F. oxysporum genotypes, it is reasonable to expect variation in the levels of endophytic aggression. Therefore, it is possible that the progenitors of VCG 01111 and VCG 01112 progenitors were predisposed to be pathogenic on cotton, and have only increased in frequency and aggressiveness as cotton has been grown extensively in Australia. Thus strictly nonpathogenic VCG 01111 and VCG 01112 progenitors may never have been present. The potential for the evolution of increased virulence in weakly pathogenic lineage A strains from native cotton hosts has recently been demonstrated experimentally (Wang et al. 2008).
Regardless of evolutionary origins, the observation that Fov strains in cultivated field soils were overwhelmingly outnumbered by nonpathogenic (on cotton) lineage E isolates was surprising for two reasons. Only one lineage E isolate was recovered from the refuge soil and this lineage accounted for only 8% of the isolates from soils associated with wild Gossypium (Table 5; Fig. 4), and based on the phylogenetic analyses, lineage E isolates are more closely related to Fov race 1–8 than are any of the pathogenic or nonpathogenic lineage A isolates (Figs 6 and 7). So despite the fact that lineage E related genotypes have become pathogenic on cotton elsewhere in the world and can increase in frequency under cultivation, in Australia the phylogenetically distinct lineage A has given rise to a new group of cotton pathogens.
Why lineage E isolates are over represented in cultivated fields is not clear, but it would appear some selective mechanism is operating. One possibility is that some lineage E isolates are pathogenic on rotation crops. Wheat has been grown in these fields in rotation with cotton and the cultivation of wheat can select for certain fungal genotypes (Edel et al. 1997). Previous studies have also demonstrated that the composition of F. oxysporum populations is affected by the application of certain fertilizers (Wang et al. 1999), and it is possible that lineage E isolates may have a fitness advantage under cultivated conditions, i.e. the application of fertilizers and the incorporation of crop debris.
The genetic structure of F. oxysporum populations in uncultivated soils and how immigration from cultivated fields impinges on these native populations and vice-versa, is largely unexplored, as is generally the case for host–pathogen interactions across the agro-ecological interface (Burdon and Thrall 2008). Gordon et al. (1992) found no spatial structure among isolates from adjacent cultivated and native California soils, with most mtDNA haplotypes occurring in both soils, indicating a high level of gene flow. In contrast, our results showed that the composition of nonpathogenic (on cotton) F. oxysporum populations from agricultural fields and uncultivated refuges differed dramatically (Table 5; Fig. 4). A better knowledge of the ecological processes underlying this dramatic shift in the composition of F. oxysporum populations will improve our understanding of the emergence of the Fusarium complex in cotton growing areas in Australia, and ultimately be useful in the development of novel control strategies and improved disease management protocols (Burdon and Thrall 2008).
This study extends our knowledge of indigenous F. oxysporum populations in Australia, but also raises interesting questions regarding the relationship of lineage A to E to other F. oxysporum and Fusarium species. The EF-1α gene sequences were compared with those in the public database using BLAST searches. The results showed that VCG 01111 and VCG 01112 of Fov found in the cotton fields as well as lineages A and E from soil associated with wild Gossypium in Australia are clearly included in the F. oxysporum clade; lineage B belongs in the Gibberella fujikuroi complex; while lineages C and D are distinct from known sequences (Fig. 7). These results reflect: (i) a lack of a one-to-one correlation between morphological, biological species, and phylogenetic species as shown in many other studies (O'Donnell et al. 1998a; Leslie et al. 2001); (ii) morphological and/or biological species show global geographic ranges, but phylogenetic species usually harbour several to many endemic species (Taylor et al. 2006); and (iii) limitations in morphological identification of F. oxysporum. The unique phylogenetic status of the two Australian VCGs of F. oxysporum f. sp. vasinfectum and lineage A in the Fusarium species complex has been recently confirmed by O'Donnell et al. (2009). Their work also showed that, within the Fusarium oxysporum species complex, only two from New Zealand showed some similarity to the Australian Fov isolates, suggesting that lineage A may be geographically restricted to the Pacific region.
Knowledge of F. oxysporum in natural ecosystems is limited although it is a common inhabitant of various native soils (McMullen and Stack 1983; Gordon et al. 1992; Summerell et al. 1993; Wang et al. 2004). However, even less is known about the extent to which pathogenic F. oxysporum strains are associated with wild relatives of a crop host and their pathogenicity towards the crop. The patterns of pathogenicity likely to be found in plant–pathogen associations are markedly affected by a range of life-history characters of the pathogen that can ultimately influence its transmission rate (Burdon 1987). It has been suggested that systems in which the pathogen is capable of saprophytic growth or is able to infect multiple host species may favour isolates that are less aggressive as transmission opportunities may be greater than for more specialized pathogens (Alexander 1981; May and Anderson 1983; Gordon and Martyn 1997). Factors such as host density and crop rotation, as well as pathogen saprophytic ability have been shown theoretically to influence the dynamics and persistence of soil-borne fungi (Thrall et al. 1997); the role of agronomic management in influencing the evolutionary trajectories of soil pathogen populations has not been widely explored.
In this study, although 15% of F. oxysporum isolates derived from soil associated with wild Gossypium showed pathogenicity on cotton (i.e. they were wild Fov) (Table 5), none killed any of the inoculated plants during the experimental period. Within the native Australian Gossypium populations, genotypes exist that are tolerant or resistant to the Fov occurring in cotton fields, while others are highly susceptible (Becerra Lopez-Lavalle et al. 2007). As a result, the selection pressures exerted by resistance differences in co-occurring wild Fov populations may have favoured the selective accumulation of isolates with enhanced pathogenicity – some of which may be pathogenic to cultivated cotton. This possibility is supported by evidence involving the occurrence of a distinct form of F. oxysporum f. sp. cubense that posed a serious economic risk to banana production in Sumatra only a few years after its establishment in the area. Tests showed that this form of F. oxysporum f. sp. cubense was asymptomatically associated with local wild bananas from which it presumably spread (Moore et al. 2001).
Wild relatives of cultivated crops have long been recognized as sources of valuable genes in resistance breeding (Kaiser et al. 1994; Bayaa et al. 1995; Huang and Lindhout 1997). However, relatively little is known about their importance either in the maintenance of the pathogens involved or in their epidemiology (Burdon and Thrall 2008). Pathogenic strains of F. oxysporum appear to gain or retain pathogenicity at the cost of losing some of their ecological breadth (Gordon and Martyn 1997). As a consequence, they risk being out-competed by nonpathogenic strains if the benefit of pathogenesis cannot be achieved regularly (Gordon and Martyn 1997). Wild Fov occurred in 61% of G. sturtianum populations, but in only a third of populations of the other three Gossypium species, suggesting a strong preference for G. sturtianum by wild Fov (Table 1). This suggests that some native Gossypium populations may not only be an inoculum reservoir for the pathogen but could also nurture the pathogen's evolutionary potential. Wild Fov occurring in all the five lineages identified in this study (Table 5) possesses significantly greater genetic diversity than does the Fov found in cotton fields that contains only two genotypes (Bentley et al. 2000). Given the proximity of cotton fields to some of these native F. oxysporum populations (e.g. in Theodore, Queensland some G. sturtianum populations occur within 200 meters of commercial cotton fields), there is little doubt that wild Fov could invade cotton fields as a result of clearing for new plantings or by dispersal in soil attached to stock or machinery.
Acknowledgments
This research was supported by the Cotton Research and Development Corporation of Australia. We thank S. Allen (Cotton Seed Distributors), D. Nehl (New South Wales Department of Primary Industries), W. Tate, B. Matheson, A. Becerra, F. Zich, J. Grace, M. Woods and J. Scown (CSIRO Plant Industry), and A. Fraser for their assistance in sample collection and experimental work.
Literature cited
- Alexander M. Why microbial predators and parasites do not eliminate their prey and host. Annual Review of Microbiology. 1981;35:113–133. doi: 10.1146/annurev.mi.35.100181.000553. [DOI] [PubMed] [Google Scholar]
- Anderson PK, Cunningham AA, Patel NG, Morales FJ, Epstein PR. Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends in Ecology and Evolution. 2004;19:535–544. doi: 10.1016/j.tree.2004.07.021. [DOI] [PubMed] [Google Scholar]
- Appel DJ, Gordon TR. Local and regional variation in populations of Fusarium oxysporum from agricultural field soils. Phytopathology. 1994;84:786–791. [Google Scholar]
- Appel DJ, Gordon TR. Relationships among pathogenic and nonpathogenic isolates of Fusarium oxysporum based on the partial sequence of the intergenic spacer region of the ribosomal DNA. Molecular Plant-Microbe Interactions. 1996;9:125–138. doi: 10.1094/mpmi-9-0125. [DOI] [PubMed] [Google Scholar]
- Armstrong GM, Armstrong JK. Formae speciales and races of Fusarium oxysporum causing wilt diseases. In: Nelson PE, Toussoun TA, Cook RJ, editors. Fusarium: Disease, Biology, and Taxonomy. University Park, PA: The Pennsylvania State University Press; 1981. pp. 391–399. [Google Scholar]
- Bayaa B, Erskine W, Hamdi A. Evaluation of a wild lentil collection for resistance to vascular wilt. Genetic Resources & Crop Evolution. 1995;42:231–235. [Google Scholar]
- Becerra Lopez-Lavalle LA, McFadden H, Brubaker CL. The effect of Gossypium C-genome chromosomes on resistance to fusarium wilt in allotetraploid cotton. Theoretical and Applied Genetics. 2007;115:477–488. doi: 10.1007/s00122-007-0581-6. [DOI] [PubMed] [Google Scholar]
- Bentley S, Kochman JK, Moore NY, Pattemore JA, Gulino L, O'Neill WT. Proceedings of the 10th Australian Cotton Conference. Brisbane, Australia: 2000. DNA diagnostics for fusarium wilt of cotton; pp. 455–461. August 16–18, 2000. [Google Scholar]
- Burdon JJ. Diseases and Plant Population Biology. Cambridge: Cambridge University Press; 1987. [Google Scholar]
- Burdon JJ, Thrall PH. Pathogen evolution across the agro-ecological interface: implications for management. Evolutionary Applications. 2008;1:57–65. doi: 10.1111/j.1752-4571.2007.00005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon JJ, Oates JD, Marshall DR. Interactions between Avena and Puccinia species. I. The wild hosts: Avena barbata Pott ex Link, A. fatua L. and A. ludoviciana Durieu. Journal of Applied Ecology. 1983;20:571–585. [Google Scholar]
- Burgess LW, Summerell BA, Bullock S, Gott KP, Backhouse D. Laboratory Manual for Fusarium Research. 3rd edn. Sydney, Australia: University of Sydney; 1994. [Google Scholar]
- Cai G, Gale LR, Schneider RW, Kistler HC, Davis RM, Elias KS, Miyao EM. Origin of race 3 of Fusarium oxysporum f. sp. lycopersici at a single site in California. Phytopathology. 2003;93:1014–1022. doi: 10.1094/PHYTO.2003.93.8.1014. [DOI] [PubMed] [Google Scholar]
- Chen Q, Ji X, Sun W. Identification of races of cotton wilt Fusarium in China. Agricultural Sciences in China. 1985;6:1–6. [Google Scholar]
- Craven LA, Stewart JMcD, Brown AHD, Grace JP. The Australian wild species of Gossypium. In: Constable GA, Forrester NW, editors. Challenging the Future: Proceedings of the World Cotton Research Conference I. Melbourne, Australia: CSIRO; 1994. pp. 278–281. [Google Scholar]
- Davis RD, Moore NY, Kochman JK. Characterisation of a population of Fusarium oxysporum f. sp. vasinfectum causing wilt of cotton in Australia. Australian Journal of Agricultural Research. 1996;47:1143–1156. [Google Scholar]
- Edel V, Steinberg C, Gautheron N, Alabouvette C. Populations of nonpathogenic Fusarium oxysporum associated with roots of four plant species compared to soilborne populations. Phytopathology. 1997;87:693–697. doi: 10.1094/PHYTO.1997.87.7.693. [DOI] [PubMed] [Google Scholar]
- Elias KS, Schneider RW, Lear MM. Analysis of vegetative compatibility groups in nonpathogenic populations of Fusarium oxysporum isolated from symptomless tomato roots. Canadian Journal of Botany. 1991;69:2089–2094. [Google Scholar]
- Ewald PW. Evolution of Infectious Diseases. Oxford: Oxford University Press; 1994. [Google Scholar]
- Fernandez D, Assigbetse K, Dubois MP, Geiger JP. Molecular characterization of races and vegetative compatibility groups in Fusarium oxysporum f. sp. vasinfectum. Applied and Environmental Microbiology. 1994;60:4039–4046. doi: 10.1128/aem.60.11.4039-4046.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel O, Shtienberg D, Abbo D, Sherman D. Sympatric ascochyta complex on wild Cicer judaicum and domesticated chickpea. Plant Pathology. 2007;56:464–471. [Google Scholar]
- Gilligan CA. Sustainable agriculture and plant diseases: an epidemiological perspective. Philosophical Transactions of the Royal Society B. 2008;363:741–759. doi: 10.1098/rstb.2007.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon TR, Martyn RD. The evolutionary biology of Fusarium oxysporum. Annual Review of Phytopathology. 1997;35:111–128. doi: 10.1146/annurev.phyto.35.1.111. [DOI] [PubMed] [Google Scholar]
- Gordon TR, Okamoto D. Population structure and the relationship between pathogenic and nonpathogenic strains of Fusarium oxysporum. Phytopathology. 1992a;82:73–77. [Google Scholar]
- Gordon TR, Okamoto D. Variation within and between populations of Fusarium oxysporum based on vegetative compatibility and mitochondrial DNA. Canadian Journal of Botany. 1992b;70:1211–1217. [Google Scholar]
- Gordon TR, Okamoto D, Milgroom MG. The structure and interrelationship of fungal populations in native and cultivated soils. Molecular Ecology. 1992;1:241–249. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hillocks RJ. Fusarium wilt. In: Hillocks RJ, editor. Cotton Diseases. Wallingford, UK: CAB International; 1992. pp. 127–160. [Google Scholar]
- Huang C, Lindhout P. Screening for resistance in wild Lycopersicon species to Fusarium oxysporum f. sp. lycopersici race 1 and race 2. Euphytica. 1997;93:145–153. [Google Scholar]
- Jeger MJ, Seal SE, Van den Bosch F. Evolutionary epidemiology of plant virus disease. Advances in Virus Research. 2006;67:163–203. doi: 10.1016/S0065-3527(06)67005-X. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Alcala-Jimenez AR, Hervas-Vargas A, Trapero-Casas JL, Jimenez-Diaz RM. Screening of wild Cicer species for resistance to races 0 and 5 of Fusarium oxysporum f. sp. ciceris. Plant Disease. 1994;78:962–967. [Google Scholar]
- Katan T, Katan J. Vegetative-compatibility grouping of Fusarium oxysporum f. sp. vasinfectum from tissue and the rhizosphere of cotton plants. Phytopathology. 1988;78:852–855. [Google Scholar]
- Katan T, Katan J, Gordon TR, Pozniak D. Physiologic races and vegetative compatibility groups of Fusarium oxysporum f. sp. melonis in Israel. Phytopathology. 1994;84:153–157. [Google Scholar]
- Kim Y, Hutmacher RB, Davis RM. Characterization of California isolates of Fusarium oxysporum f. sp. vasinfectum. Plant Disease. 2005;89:366–372. doi: 10.1094/PD-89-0366. [DOI] [PubMed] [Google Scholar]
- Kochman JK. Fusarium wilt in cotton – a new record in Australia. Australasian Plant Pathology. 1995;24:74. [Google Scholar]
- Leslie JF, Summerell BA. The Fusarium Laboratory Manual. Ames, IA: Blackwell Publishing; 2006. [Google Scholar]
- Leslie JF, Zeller KA, Summerell BA. Icebergs and species in populations of Fusarium. Physiological and Molecular Plant Pathology. 2001;59:107–117. [Google Scholar]
- May RM, Anderson RM. Epidemiology and genetics in the coevolution of parasites and hosts. Proceedings of the Royal Society London B. 1983;219:281–313. doi: 10.1098/rspb.1983.0075. [DOI] [PubMed] [Google Scholar]
- McMullen MP, Stack RW. Fusarium species associated with grassland soils. Canadian Journal of Botany. 1983;61:2530–2538. [Google Scholar]
- Moore NY, Pegg KG, Buddenhagen IW, Bentley S. Fusarium wilt of banana: a diverse clonal pathogen of a domesticated clonal host. In: Summerell BA, Leslie JF, Backhouse D, Bryden WL, Burgess LW, editors. Fusarium: Paul E. Nelson Memorial Symposium. St Paul, MN: APS Press; 2001. pp. 212–224. [Google Scholar]
- Oates JD, Burdon JJ, Brouwer JB. Interactions between Avena and Puccinia species. II. The pathogens: Puccinia coronata Cda and P. graminis Pers. f.sp. avenae Eriks. and Henn. Journal of Applied Ecology. 1983;20:585–596. [Google Scholar]
- O'Donnell K, Cigelnik E, Nirenberg HI. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia. 1998a;90:465–493. [Google Scholar]
- O'Donnell K, Kistler HC, Cigelnik E, Ploetz RC. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences USA. 1998b;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K, Kistler HC, Tacke BK, Casper HH. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proceedings of the National Academy of Sciences USA. 2000;97:7905–7910. doi: 10.1073/pnas.130193297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K, Gueidan C, Sink S, Johnston PR, Crous PW, Glenn A, Riley R, et al. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genetics and Biology. 2009;46:936–948. doi: 10.1016/j.fgb.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Comparative Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Puhalla JE. Classification of strains of Fusarium oxysporum on the basis of vegetative compatibility. Canadian Journal of Botany. 1985;63:179–183. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rosewich UL, Pettway RE, Katan T, Kistler HC. Population genetic analysis corroborates dispersal of Fusarium oxysporum f. sp. radicis lycopersici from Florida to Europe. Phytopathology. 1999;89:623–630. doi: 10.1094/PHYTO.1999.89.8.623. [DOI] [PubMed] [Google Scholar]
- Skovgaard K, Nirenberg HI, O'Donnell K, Rosendahl S. Evolution of Fusarium oxysporum f. sp. vasinfectum races inferred from multigene genealogies. Phytopathology. 2001;91:1231–1237. doi: 10.1094/PHYTO.2001.91.12.1231. [DOI] [PubMed] [Google Scholar]
- Skovgaard K, Bodker L, Rosendahl S. Population structure and pathogenicity of members of the Fusarium oxysporum complex isolated from soil and root necrosis of pea (Pisum sativum L.) FEMS Microbiology Ecology. 2002;42:367–374. doi: 10.1111/j.1574-6941.2002.tb01026.x. [DOI] [PubMed] [Google Scholar]
- Summerell BA, Rugg CA, Burgess LW. Mycogeography of Fusarium: survey of Fusarium species associated with forest and woodland communities in north Queensland, Australia. Mycological Research. 1993;97:1015–1019. [Google Scholar]
- Taylor JW, Turner E, Townsend JP, Dettman JR, Jacobson D. Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom Fungi. Philosophical Transactions of the Royal Society B. 2006;361:1947–1963. doi: 10.1098/rstb.2006.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JN. The Geographic Mosaic of Coevolution. Chicago, IL: University of Chicago Press; 2005. [Google Scholar]
- Thrall PH, Bever JD, Mihail JD, Alexander HM. The population dynamics of annual plants and soil-borne fungal pathogens. Journal of Ecology. 1997;85:313–328. [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, Van De Lee T, Hornes M, Frijters A, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Dale ML, Kochman JK, Allen SJ, Obst NR. Variations in soil populations of Fusarium oxysporum f. sp. vasinfectum as influenced by fertiliser application and growth of different crops. Australasian Plant Pathology. 1999;28:174–181. [Google Scholar]
- Wang B, Brubaker CL, Burdon JJ. Fusarium species and Fusarium wilt pathogens associated with native Gossypium populations in Australia. Mycological Research. 2004;108:35–44. doi: 10.1017/s0953756203008803. [DOI] [PubMed] [Google Scholar]
- Wang B, Brubaker CL, Tate W, Woods MJ, Matheson BA, Burdon JJ. Genetic variation and population structure of Fusarium oxysporum f. sp. vasinfectum in Australia. Plant Pathology. 2006;55:746–755. [Google Scholar]
- Wang B, Brubaker CL, Tate W, Woods MJ, Burdon JJ. Evolution of virulence in Fusarium oxysporum f. sp. vasinfectum using serial passage assays through susceptible cotton. Phytopathology. 2008;98:296–303. doi: 10.1094/PHYTO-98-3-0296. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innes MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. San Diego, CA: Academic Press; 1990. pp. 315–322. [Google Scholar]
- Woudt LP, Neuvel A, Sikkema A, Van Grinswen MQZM, De Milliano WAJ. Genetic variation in Fusarium oxysporum from cyclamen. Phytopathology. 1995;85:1348–1355. [Google Scholar]


