Figure 5.
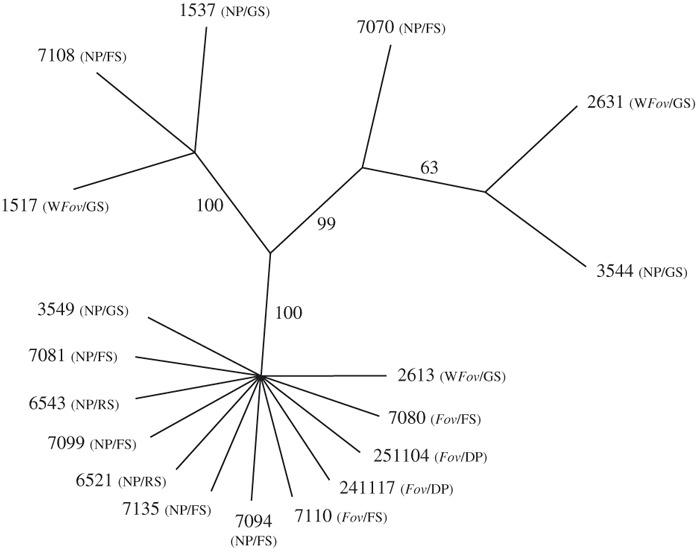
The single most parsimonious unrooted topology (length = 75 steps; consistency index = 0.9867; retention index = 0.9939) obtained from a heuristic parsimony optimized analysis of a concatenated matrix of the translation elongation factor-1α (EF-1α) gene and mitochondrial small subunit (mtSSU) rDNA sequences from four Fov, eight lineage A, three lineage B, and three lineage E isolates. Bootstrap values (10 000 replicates) are placed beside each branch of the typology. The pathogenicity of isolates on cotton (before slash; Fov = F. oxysporum f. sp. vasinfectum; WFov = wild F. oxysporum f. sp. vasinfectum; NP = nonpathogenic) and their origin (behind slash; GS = Gossypium soil; RS = uncultivated refuge soil; FS = cultivated field soil; DP = diseased plant) are given in brackets.
